Fungal Ku prevents permanent cell cycle arrest by suppressing DNA damage signaling at telomeres
- PMID: 25653166
- PMCID: PMC4344518
- DOI: 10.1093/nar/gkv082
Fungal Ku prevents permanent cell cycle arrest by suppressing DNA damage signaling at telomeres
Abstract
The Ku heterodimer serves in the initial step in repairing DNA double-strand breaks by the non-homologous end-joining pathway. Besides this key function, Ku also plays a role in other cellular processes including telomere maintenance. Inactivation of Ku can lead to DNA repair defects and telomere aberrations. In model organisms where Ku has been studied, inactivation can lead to DNA repair defects and telomere aberrations. In general Ku deficient mutants are viable, but a notable exception to this is human where Ku has been found to be essential. Here we report that similar to the situation in human Ku is required for cell proliferation in the fungus Ustilago maydis. Using conditional strains for Ku expression, we found that cells arrest permanently in G2 phase when Ku expression is turned off. Arrest results from cell cycle checkpoint activation due to persistent signaling via the DNA damage response (DDR). Our results point to the telomeres as the most likely source of the DNA damage signal. Inactivation of the DDR makes the Ku complex dispensable for proliferation in this organism. Our findings suggest that in U. maydis, unprotected telomeres arising from Ku depletion are the source of the signal that activates the DDR leading to cell cycle arrest.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
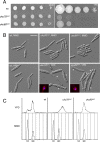
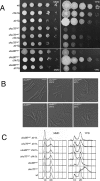
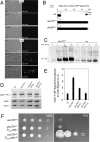


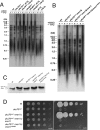
References
-
- Deriano L., Roth D.B. Modernizing the nonhomologous end-joining repertoire: alternative and classical NHEJ share the stage. Annu. Rev. Genet. 2013;47:433–455. - PubMed
-
- Panier S., Boulton S.J. Double-strand break repair: 53BP1 comes into focus. Nat. Rev. Mol. Cell Biol. 2014;15:7–18. - PubMed
-
- Downs J.A., Jackson S.P. A means to a DNA end: the many roles of Ku. Nat. Rev. Mol. Cell Biol. 2004;5:367–378. - PubMed

