Yeast Kre33 and human NAT10 are conserved 18S rRNA cytosine acetyltransferases that modify tRNAs assisted by the adaptor Tan1/THUMPD1
- PMID: 25653167
- PMCID: PMC4344512
- DOI: 10.1093/nar/gkv075
Yeast Kre33 and human NAT10 are conserved 18S rRNA cytosine acetyltransferases that modify tRNAs assisted by the adaptor Tan1/THUMPD1
Abstract
The function of RNA is subtly modulated by post-transcriptional modifications. Here, we report an important crosstalk in the covalent modification of two classes of RNAs. We demonstrate that yeast Kre33 and human NAT10 are RNA cytosine acetyltransferases with, surprisingly, specificity toward both 18S rRNA and tRNAs. tRNA acetylation requires the intervention of a specific and conserved adaptor: yeast Tan1/human THUMPD1. In budding and fission yeasts, and in human cells, we found two acetylated cytosines on 18S rRNA, one in helix 34 important for translation accuracy and another in helix 45 near the decoding site. Efficient 18S rRNA acetylation in helix 45 involves, in human cells, the vertebrate-specific box C/D snoRNA U13, which, we suggest, exposes the substrate cytosine to modification through Watson-Crick base pairing with 18S rRNA precursors during small subunit biogenesis. Finally, while Kre33 and NAT10 are essential for pre-rRNA processing reactions leading to 18S rRNA synthesis, we demonstrate that rRNA acetylation is dispensable to yeast cells growth. The inactivation of NAT10 was suggested to suppress nuclear morphological defects observed in laminopathic patient cells through loss of microtubules modification and cytoskeleton reorganization. We rather propose the effects of NAT10 on laminopathic cells are due to reduced ribosome biogenesis or function.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

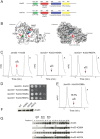


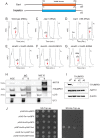
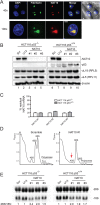

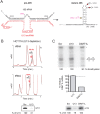
References
-
- Motorin Y., Helm M. RNA nucleotide methylation. Wiley Interdiscip. Rev. RNA. 2011;2:611–631. - PubMed
-
- Grosjean H. Nucleic acids are not boring long polymers of only four types of nucleotides: a guided tour. In: Grosjean H, editor. DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution. Austin, TX: Landes Bioscience; 2009. pp. 1–18.
-
- Decatur W.A., Fournier M.J. rRNA modifications and ribosome function. Trends Biochem. Sci. 2002;27:344–351. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

