Emergence of an outbreak-associated Clostridium difficile variant with increased virulence
- PMID: 25653402
- PMCID: PMC4365207
- DOI: 10.1128/JCM.03058-14
Emergence of an outbreak-associated Clostridium difficile variant with increased virulence
Abstract
The prevalence of Clostridium difficile infections has increased due to the emergence of epidemic variants from diverse genetic lineages. Here we describe the emergence of a novel variant during an outbreak in a Costa Rican hospital that was associated with severe clinical presentations. This C. difficile variant elicited higher white blood cell counts and caused disease in younger patients than did other strains isolated during the outbreak. Furthermore, it had a recurrence rate, a 30-day attributable disease rate, and disease severity as great as those of the epidemic strain NAP1. Pulsed-field gel electrophoresis genotyping indicated that the outbreak strains belong to a previously undescribed variant, designated NAPCR1. Whole-genome sequencing and ribotyping indicated that the NAPCR1 variant belongs to C. difficile ribotype 012 and sequence type 54, as does the reference strain 630. NAPCR1 strains are resistant to fluoroquinolones due to a mutation in gyrA, and they possess an 18-bp deletion in tcdC that is characteristic of the epidemic, evolutionarily distinct, C. difficile NAP1 variant. NAPCR1 genomes contain 10% more predicted genes than strain 630, most of which are of hypothetical function and are present on phages and other mobile genetic elements. The increased virulence of NAPCR1 was confirmed by mortality rates in the hamster model and strong inflammatory responses induced by bacteria-free supernatants in the murine ligated loop model. However, NAPCR1 strains do not synthesize toxin A and toxin B at levels comparable to those in NAP1 strains. Our results suggest that the pathogenic potential of this emerging C. difficile variant is due to the acquisition of hypothetical functions associated with laterally acquired DNA.
Copyright © 2015, Quesada-Gómez et al.
Figures
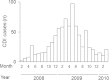
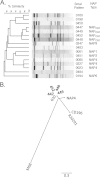
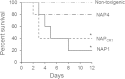
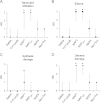
Similar articles
-
Predominance and high antibiotic resistance of the emerging Clostridium difficile genotypes NAPCR1 and NAP9 in a Costa Rican hospital over a 2-year period without outbreaks.Emerg Microbes Infect. 2016 May 11;5(5):e42. doi: 10.1038/emi.2016.38. Emerg Microbes Infect. 2016. PMID: 27165560 Free PMC article.
-
A Clostridium difficile Lineage Endemic to Costa Rican Hospitals Is Multidrug Resistant by Acquisition of Chromosomal Mutations and Novel Mobile Genetic Elements.Antimicrob Agents Chemother. 2017 Mar 24;61(4):e02054-16. doi: 10.1128/AAC.02054-16. Print 2017 Apr. Antimicrob Agents Chemother. 2017. PMID: 28137804 Free PMC article.
-
Lack of association between clinical outcome of Clostridium difficile infections, strain type, and virulence-associated phenotypes.J Clin Microbiol. 2011 Dec;49(12):4040-6. doi: 10.1128/JCM.05053-11. Epub 2011 Sep 28. J Clin Microbiol. 2011. PMID: 21956985 Free PMC article.
-
Clostridium difficile infection: toxins and non-toxin virulence factors, and their contributions to disease establishment and host response.Gut Microbes. 2012 Mar-Apr;3(2):121-34. doi: 10.4161/gmic.19399. Epub 2012 Mar 1. Gut Microbes. 2012. PMID: 22555464 Free PMC article. Review.
-
The changing epidemiology of Clostridium difficile infections.Clin Microbiol Rev. 2010 Jul;23(3):529-49. doi: 10.1128/CMR.00082-09. Clin Microbiol Rev. 2010. PMID: 20610822 Free PMC article. Review.
Cited by
-
Mechanisms underpinning the efficacy of faecal microbiota transplantation in treating gastrointestinal disease.Therap Adv Gastroenterol. 2020 Sep 3;13:1756284820946904. doi: 10.1177/1756284820946904. eCollection 2020. Therap Adv Gastroenterol. 2020. PMID: 32952613 Free PMC article. Review.
-
Stable core virome despite variable microbiome after fecal transfer.Gut Microbes. 2017 May 4;8(3):214-220. doi: 10.1080/19490976.2016.1265196. Epub 2016 Dec 9. Gut Microbes. 2017. PMID: 27935413 Free PMC article. Review.
-
Analysis of TcdB Proteins within the Hypervirulent Clade 2 Reveals an Impact of RhoA Glucosylation on Clostridium difficile Proinflammatory Activities.Infect Immun. 2016 Jan 11;84(3):856-65. doi: 10.1128/IAI.01291-15. Infect Immun. 2016. PMID: 26755157 Free PMC article.
-
Burden of Clostridium (Clostridioides) difficile Infection among Patients in Western Asia: A Systematic Review and Meta-Analysis.Iran J Public Health. 2019 Sep;48(9):1589-1599. Iran J Public Health. 2019. PMID: 31700814 Free PMC article. Review.
-
The Global Burden of Clostridioides difficile Infections, 2016-2024: A Systematic Review and Meta-Analysis.Infect Dis Rep. 2025 Apr 14;17(2):31. doi: 10.3390/idr17020031. Infect Dis Rep. 2025. PMID: 40277958 Free PMC article. Review.
References
-
- Killgore G, Thompson A, Johnson S, Brazier J, Kuijper E, Pepin J, Frost EH, Savelkoul P, Nicholson B, van den Berg RJ, Kato H, Sambol SP, Zukowski W, Woods C, Limbago B, Gerding DN, McDonald LC. 2008. Comparison of seven techniques for typing international epidemic strains of Clostridium difficile: restriction endonuclease analysis, pulsed-field gel electrophoresis, PCR-ribotyping, multilocus sequence typing, multilocus variable-number tandem-repeat analysis, amplified fragment length polymorphism, and surface layer protein A gene sequence typing. J Clin Microbiol 46:431–437. doi:10.1128/JCM.01484-07. - DOI - PMC - PubMed
-
- Smith A. 2005. Outbreak of Clostridium difficile infection in an English hospital linked to hypertoxin-producing strains in Canada and the US. Euro Surveill 10(26):pii=2735 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=2735. - PubMed
Publication types
MeSH terms
Substances
Associated data
- BioProject/PRJNA264745
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

