Fine mapping and characterization of the L-polymerase-binding domain of the respiratory syncytial virus phosphoprotein
- PMID: 25653447
- PMCID: PMC4442346
- DOI: 10.1128/JVI.03619-14
Fine mapping and characterization of the L-polymerase-binding domain of the respiratory syncytial virus phosphoprotein
Abstract
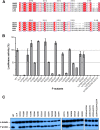
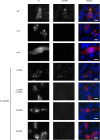
The minimum requirement for an active RNA-dependent RNA polymerase of respiratory syncytial virus (RSV) is a complex made of two viral proteins, the polymerase large protein (L) and the phosphoprotein (P). Here we have investigated the domain on P that is responsible for this critical P-L interaction. By use of recombinant proteins and serial deletions, an L binding site was mapped in the C-terminal region of P, just upstream of the N-RNA binding site. The role of this molecular recognition element of about 30 amino acid residues in the L-P interaction and RNA polymerase activity was evaluated in cellula using an RSV minigenome system and site-directed mutagenesis. The results highlighted the critical role of hydrophobic residues located in this region.
Importance: Respiratory syncytial virus (RSV) is the leading cause of lower respiratory tract illness in infants. Since no vaccine and no good antivirals against RSV are available, it is essential to better understand how the viral machinery functions in order to develop new antiviral strategies. Like all negative-strand RNA viruses, RSV codes for its own machinery to replicate and transcribe its genome. The core of this machinery is composed of two proteins, the phosphoprotein (P) and the large protein (L). Here, using recombinant proteins, we have mapped and characterized the P domain responsible for this L-P interaction and the formation of an active L-P complex. These findings extend our understanding of the mechanism of action of RSV RNA polymerase and allow us to define a new target for the development of drugs against RSV.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures





Similar articles
-
Identification and characterization of the binding site of the respiratory syncytial virus phosphoprotein to RNA-free nucleoprotein.J Virol. 2015 Apr;89(7):3484-96. doi: 10.1128/JVI.03666-14. Epub 2015 Jan 7. J Virol. 2015. PMID: 25568210 Free PMC article.
-
A Druggable Pocket at the Nucleocapsid/Phosphoprotein Interaction Site of Human Respiratory Syncytial Virus.J Virol. 2015 Nov;89(21):11129-43. doi: 10.1128/JVI.01612-15. Epub 2015 Aug 5. J Virol. 2015. PMID: 26246564 Free PMC article.
-
In Vitro Primer-Based RNA Elongation and Promoter Fine Mapping of the Respiratory Syncytial Virus.J Virol. 2020 Dec 9;95(1):e01897-20. doi: 10.1128/JVI.01897-20. Print 2020 Dec 9. J Virol. 2020. PMID: 33028717 Free PMC article.
-
New antiviral approaches for respiratory syncytial virus and other mononegaviruses: Inhibiting the RNA polymerase.Antiviral Res. 2016 Oct;134:63-76. doi: 10.1016/j.antiviral.2016.08.006. Epub 2016 Aug 27. Antiviral Res. 2016. PMID: 27575793 Review.
-
Biochemistry of the Respiratory Syncytial Virus L Protein Embedding RNA Polymerase and Capping Activities.Viruses. 2023 Jan 25;15(2):341. doi: 10.3390/v15020341. Viruses. 2023. PMID: 36851554 Free PMC article. Review.
Cited by
-
Discovery of a non-nucleoside inhibitor that binds to a novel site in the palm domain of the respiratory syncytial virus RNA-dependent RNA polymerase.J Virol. 2025 Jul 22;99(7):e0017825. doi: 10.1128/jvi.00178-25. Epub 2025 Jun 2. J Virol. 2025. PMID: 40454903 Free PMC article.
-
Host CARD11 Inhibits Newcastle Disease Virus Replication by Suppressing Viral Polymerase Activity in Neurons.J Virol. 2019 Nov 26;93(24):e01499-19. doi: 10.1128/JVI.01499-19. Print 2019 Dec 15. J Virol. 2019. PMID: 31554683 Free PMC article.
-
Reversion mutations in phosphoprotein P of a codon-pair-deoptimized human respiratory syncytial virus confer increased transcription, immunogenicity, and genetic stability without loss of attenuation.PLoS Pathog. 2021 Dec 29;17(12):e1010191. doi: 10.1371/journal.ppat.1010191. eCollection 2021 Dec. PLoS Pathog. 2021. PMID: 34965283 Free PMC article.
-
Polymerases of paramyxoviruses and pneumoviruses.Virus Res. 2017 Apr 15;234:87-102. doi: 10.1016/j.virusres.2017.01.008. Epub 2017 Jan 16. Virus Res. 2017. PMID: 28104450 Free PMC article. Review.
-
Tetramerization of Phosphoprotein is Essential for Respiratory Syncytial Virus Budding while its N Terminal Region Mediates Direct Interactions with the Matrix Protein.J Virol. 2021 Mar 10;95(7):e02217-20. doi: 10.1128/JVI.02217-20. Epub 2021 Jan 6. J Virol. 2021. PMID: 33408180 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

