Stress granule components G3BP1 and G3BP2 play a proviral role early in Chikungunya virus replication
- PMID: 25653451
- PMCID: PMC4442398
- DOI: 10.1128/JVI.03612-14
Stress granule components G3BP1 and G3BP2 play a proviral role early in Chikungunya virus replication
Abstract
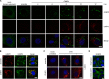
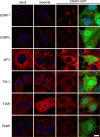
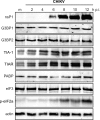
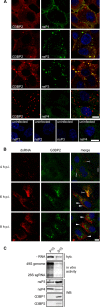
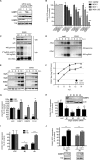
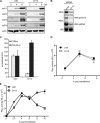
Stress granules (SGs) are protein-mRNA aggregates that are formed in response to environmental stresses, resulting in translational inhibition. SGs are generally believed to play an antiviral role and are manipulated by many viruses, including various alphaviruses. GTPase-activating protein (SH3 domain)-binding protein 1 (G3BP1) is a key component and commonly used marker of SGs. Its homolog G3BP2 is a less extensively studied SG component. Here, we demonstrate that Chikungunya virus (CHIKV) infection induces cytoplasmic G3BP1- and G3BP2-containing granules that differ from bona fide SGs in terms of morphology, composition, and behavior. For several Old World alphaviruses it has been shown that nonstructural protein 3 (nsP3) interacts with G3BPs, presumably to inhibit SG formation, and we have confirmed this interaction in CHIKV-infected cells. Surprisingly, CHIKV also relied on G3BPs for efficient replication, as simultaneous depletion of G3BP1 and G3BP2 reduced viral RNA levels, CHIKV protein expression, and viral progeny titers. The G3BPs colocalized with CHIKV nsP2 and nsP3 in cytoplasmic foci, but no colocalization with nsP1, nsP4, or dsRNA was observed. Furthermore, G3BPs could not be detected in a cellular fraction enriched for CHIKV replication/transcription complexes, suggesting that they are not directly involved in CHIKV RNA synthesis. Depletion of G3BPs did not affect viral entry, translation of incoming genomes, or nonstructural polyprotein processing but resulted in severely reduced levels of negative-stranded (and consequently also positive-stranded) RNA. This suggests a role for the G3BPs in the switch from translation to genome amplification, although the exact mechanism by which they act remains to be explored.
Importance: Chikungunya virus (CHIKV) causes a severe polyarthritis that has affected millions of people since its reemergence in 2004. The lack of approved vaccines or therapeutic options and the ongoing explosive outbreak in the Caribbean underline the importance of better understanding CHIKV replication. Stress granules (SGs) are cytoplasmic protein-mRNA aggregates formed in response to various stresses, including viral infection. The RNA-binding proteins G3BP1 and G3BP2 are essential SG components. SG formation and the resulting translational inhibition are generally considered an antiviral response, and many viruses manipulate or block this process. Late in infection, we and others have observed CHIKV nonstructural protein 3 in cytoplasmic G3BP1- and G3BP2-containing granules. These virally induced foci differed from true SGs and did not appear to represent replication complexes. Surprisingly, we found that G3BP1 and G3BP2 were also needed for efficient CHIKV replication, likely by facilitating the switch from translation to genome amplification early in infection.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures

Similar articles
-
The SHFV nsp2 and nucleocapsid proteins recruit G3BP1 to sites of viral replication, but stress granules are not induced by the infection.J Virol. 2025 Jul 22;99(7):e0079425. doi: 10.1128/jvi.00794-25. Epub 2025 Jun 23. J Virol. 2025. PMID: 40548742 Free PMC article.
-
Separate domains of G3BP promote efficient clustering of alphavirus replication complexes and recruitment of the translation initiation machinery.PLoS Pathog. 2019 Jun 14;15(6):e1007842. doi: 10.1371/journal.ppat.1007842. eCollection 2019 Jun. PLoS Pathog. 2019. PMID: 31199850 Free PMC article.
-
Pseudorabies virus IE180 protein hijacks G3BPs into the nucleus to inhibit stress granule formation.J Virol. 2025 Apr 15;99(4):e0208824. doi: 10.1128/jvi.02088-24. Epub 2025 Mar 27. J Virol. 2025. PMID: 40145738 Free PMC article.
-
Two Birds With One Stone: RNA Virus Strategies to Manipulate G3BP1 and Other Stress Granule Components.Wiley Interdiscip Rev RNA. 2025 Mar-Apr;16(2):e70005. doi: 10.1002/wrna.70005. Wiley Interdiscip Rev RNA. 2025. PMID: 40170442 Free PMC article. Review.
-
Rasputin a decade on and more promiscuous than ever? A review of G3BPs.Biochim Biophys Acta Mol Cell Res. 2019 Mar;1866(3):360-370. doi: 10.1016/j.bbamcr.2018.09.001. Epub 2018 Sep 5. Biochim Biophys Acta Mol Cell Res. 2019. PMID: 30595162 Free PMC article. Review.
Cited by
-
A proposed role for the SARS-CoV-2 nucleocapsid protein in the formation and regulation of biomolecular condensates.FASEB J. 2020 Aug;34(8):9832-9842. doi: 10.1096/fj.202001351. Epub 2020 Jun 20. FASEB J. 2020. PMID: 32562316 Free PMC article.
-
The interaction of YBX1 with G3BP1 promotes renal cell carcinoma cell metastasis via YBX1/G3BP1-SPP1- NF-κB signaling axis.J Exp Clin Cancer Res. 2019 Sep 3;38(1):386. doi: 10.1186/s13046-019-1347-0. J Exp Clin Cancer Res. 2019. PMID: 31481087 Free PMC article.
-
Pro-Viral and Anti-Viral Roles of the RNA-Binding Protein G3BP1.Viruses. 2023 Feb 6;15(2):449. doi: 10.3390/v15020449. Viruses. 2023. PMID: 36851663 Free PMC article. Review.
-
Zika Virus Subverts Stress Granules To Promote and Restrict Viral Gene Expression.J Virol. 2019 May 29;93(12):e00520-19. doi: 10.1128/JVI.00520-19. Print 2019 Jun 15. J Virol. 2019. PMID: 30944179 Free PMC article.
-
Mosquito Rasputin interacts with chikungunya virus nsP3 and determines the infection rate in Aedes albopictus.Parasit Vectors. 2015 Sep 17;8:464. doi: 10.1186/s13071-015-1070-4. Parasit Vectors. 2015. PMID: 26384002 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

