Proteomics analysis of urine reveals acute phase response proteins as candidate diagnostic biomarkers for prostate cancer
- PMID: 25653573
- PMCID: PMC4316650
- DOI: 10.1186/s12953-014-0059-9
Proteomics analysis of urine reveals acute phase response proteins as candidate diagnostic biomarkers for prostate cancer
Abstract
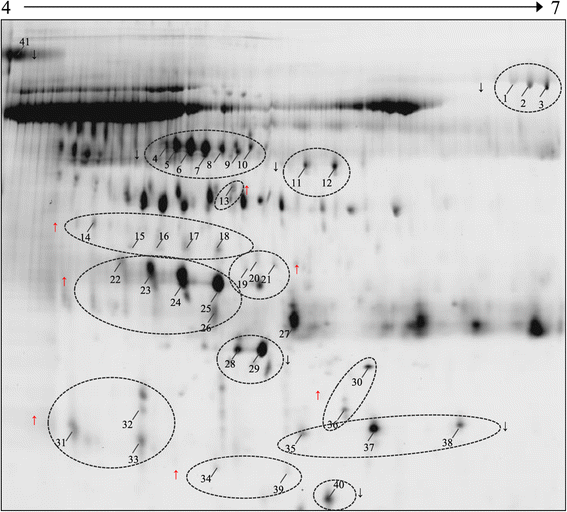
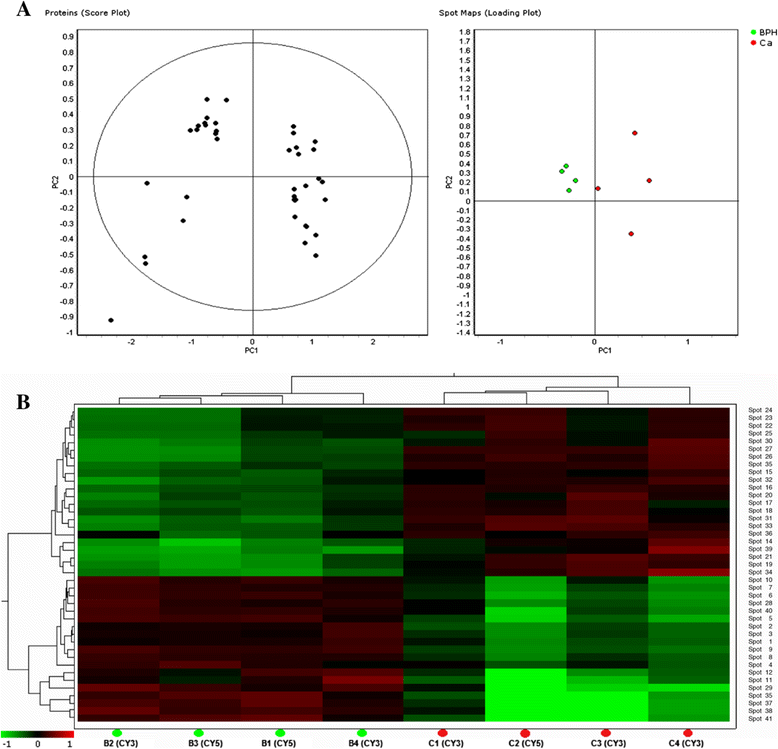
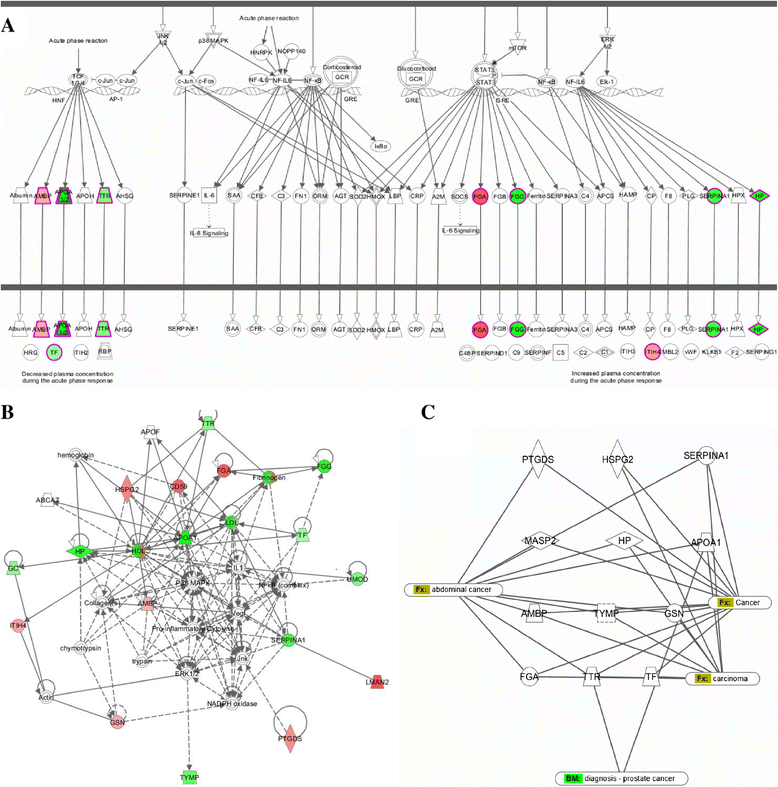
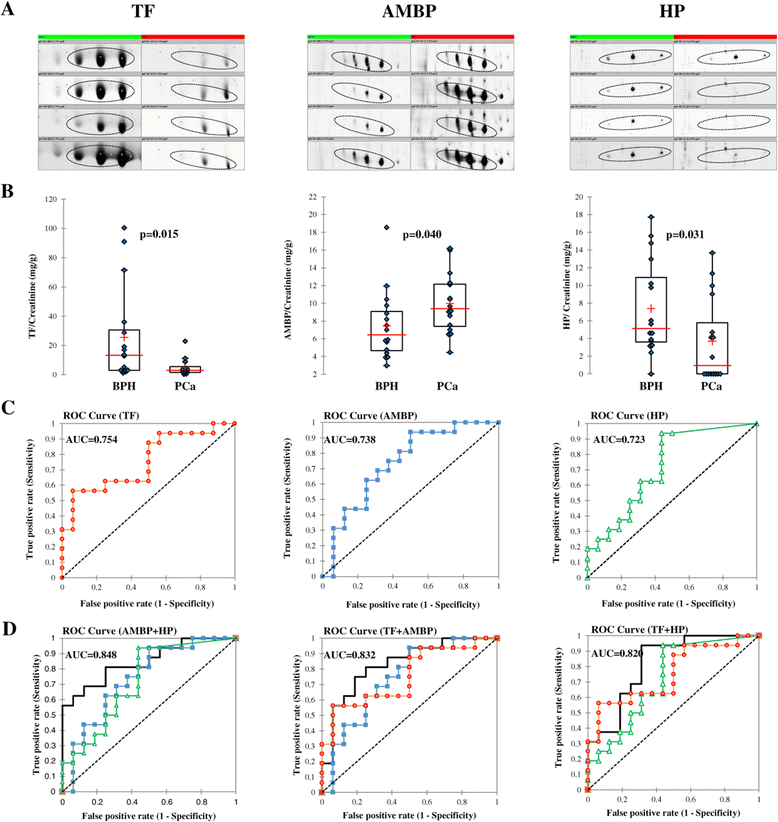
Despite the overall success of prostate specific antigen (PSA) in screening and detection of prostate cancer (PCa), its use has been limited due to the lack of specificity. The principal driving goal currently within PCa research is to identify non-invasive biomarker(s) for early detection of aggressive tumors with greater sensitivity and specificity than PSA. In this study, we focused on identification of non-invasive biomarkers in urine with higher specificity than PSA. We tested urine samples from PCa and benign prostatic hyperplasia (BPH) patients by 2-D DIGE coupled with MS and bioinformatics analysis. Statistically significant (p < 0.05), 1.8 fold variation or more in abundance, showed 41 spots, corresponding to 23 proteins. The Ingenuity Pathway Analysis showed significant association with the Acute Phase Response Signaling pathway. Nine proteins with differential abundances were included in this pathway: AMBP, APOA1, FGA, FGG, HP, ITIH4, SERPINA1, TF and TTR. The expression pattern of 4 acute phase response proteins differed from the defined expression in the canonical pathway. The urine levels of TF, AMPB and HP were measured by immunoturbidimetry in an independent validation set. The concentration of AMPB in urine was significantly higher in PCa while levels of TF and HP were opposite (p < 0.05). The AUC for the individual proteins ranged from 0.723 to 0.754. The combination of HP and AMBP yielded the highest accuracy (AUC = 0.848), greater than PSA. The proposed biomarker set is quickly quantifiable and economical with potential to improve the sensitivity and specificity of PCa detection.
Keywords: 2-D DIGE; Benign prostate hyperplasia; MS; Non-invasive biomarkers; Prostate cancer; Urine analysis.
Figures
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

