Interactions of Salmonella with animals and plants
- PMID: 25653644
- PMCID: PMC4301013
- DOI: 10.3389/fmicb.2014.00791
Interactions of Salmonella with animals and plants
Abstract
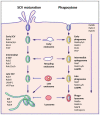
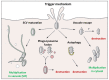
Salmonella enterica species are Gram-negative bacteria, which are responsible for a wide range of food- and water-borne diseases in both humans and animals, thereby posing a major threat to public health. Recently, there has been an increasing number of reports, linking Salmonella contaminated raw vegetables and fruits with food poisoning. Many studies have shown that an essential feature of the pathogenicity of Salmonella is its capacity to cross a number of barriers requiring invasion of a large variety of cells and that the extent of internalization may be influenced by numerous factors. However, it is poorly understood how Salmonella successfully infects hosts as diversified as animals or plants. The aim of this review is to describe the different stages required for Salmonella interaction with its hosts: (i) attachment to host surfaces; (ii) entry processes; (iii) multiplication; (iv) suppression of host defense mechanisms; and to point out similarities and differences between animal and plant infections.
Keywords: Salmonella infections; adhesion; host defense strategies; invasion mechanisms; multiplication.
Figures

Similar articles
-
Multiplicity of Salmonella entry mechanisms, a new paradigm for Salmonella pathogenesis.Microbiologyopen. 2012 Sep;1(3):243-58. doi: 10.1002/mbo3.28. Epub 2012 Jun 18. Microbiologyopen. 2012. PMID: 23170225 Free PMC article.
-
Mechanisms adopted by Salmonella to colonize plant hosts.Food Microbiol. 2021 Oct;99:103833. doi: 10.1016/j.fm.2021.103833. Epub 2021 May 20. Food Microbiol. 2021. PMID: 34119117 Review.
-
Salmonella, a cross-kingdom pathogen infecting humans and plants.FEMS Microbiol Lett. 2013 Jun;343(1):1-7. doi: 10.1111/1574-6968.12127. Epub 2013 Apr 2. FEMS Microbiol Lett. 2013. PMID: 23488473 Review.
-
An Updated View on the Rck Invasin of Salmonella: Still Much to Discover.Front Cell Infect Microbiol. 2017 Dec 8;7:500. doi: 10.3389/fcimb.2017.00500. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 29276700 Free PMC article. Review.
-
Plants as alternative hosts for Salmonella.Trends Plant Sci. 2012 May;17(5):245-9. doi: 10.1016/j.tplants.2012.03.007. Epub 2012 Apr 17. Trends Plant Sci. 2012. PMID: 22513107
Cited by
-
Transcriptomic Approach for Understanding the Adaptation of Salmonella enterica to Contaminated Produce.J Microbiol Biotechnol. 2020 Nov 28;30(11):1729-1738. doi: 10.4014/jmb.2007.07036. J Microbiol Biotechnol. 2020. PMID: 32830190 Free PMC article.
-
Role of Fimbriae, Flagella and Cellulose on the Attachment of Salmonella Typhimurium ATCC 14028 to Plant Cell Wall Models.PLoS One. 2016 Jun 29;11(6):e0158311. doi: 10.1371/journal.pone.0158311. eCollection 2016. PLoS One. 2016. PMID: 27355584 Free PMC article.
-
Epidemiology of foodborne diseases caused by Salmonella in Zhejiang Province, China, between 2010 and 2021.Front Public Health. 2023 Feb 1;11:1127925. doi: 10.3389/fpubh.2023.1127925. eCollection 2023. Front Public Health. 2023. PMID: 36817893 Free PMC article.
-
Antimicrobial mechanism of semi-bionic extracts of three traditional medicinal plants-Rheum palmatum L., Scutellaria baicalensis Georgi, and Houttuynia cordata Thunb-That can be used as antibiotic alternatives.Front Vet Sci. 2023 Jan 13;9:1083223. doi: 10.3389/fvets.2022.1083223. eCollection 2022. Front Vet Sci. 2023. PMID: 36713859 Free PMC article.
-
Microgravity and evasion of plant innate immunity by human bacterial pathogens.NPJ Microgravity. 2023 Sep 7;9(1):71. doi: 10.1038/s41526-023-00323-x. NPJ Microgravity. 2023. PMID: 37679341 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

