SATRAT: Staphylococcus aureus transcript regulatory network analysis tool
- PMID: 25653902
- PMCID: PMC4304862
- DOI: 10.7717/peerj.717
SATRAT: Staphylococcus aureus transcript regulatory network analysis tool
Abstract
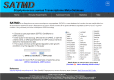
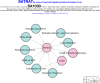
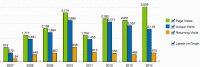
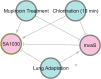
Staphylococcus aureus is a commensal organism that primarily colonizes the nose of healthy individuals. S. aureus causes a spectrum of infections that range from skin and soft-tissue infections to fatal invasive diseases. S. aureus uses a large number of virulence factors that are regulated in a coordinated fashion. The complex regulatory mechanisms have been investigated in numerous high-throughput experiments. Access to this data is critical to studying this pathogen. Previously, we developed a compilation of microarray experimental data to enable researchers to search, browse, compare, and contrast transcript profiles. We have substantially updated this database and have built a novel exploratory tool-SATRAT-the S. aureus transcript regulatory network analysis tool, based on the updated database. This tool is capable of performing deep searches using a query and generating an interactive regulatory network based on associations among the regulators of any query gene. We believe this integrated regulatory network analysis tool would help researchers explore the missing links and identify novel pathways that regulate virulence in S. aureus. Also, the data model and the network generation code used to build this resource is open sourced, enabling researchers to build similar resources for other bacterial systems.
Keywords: Infectious disease; RNA sequencing; RNA-Seq; Staphylococcus aureus; Staphylococcus aureus microarray meta-database (SAMMD); Staphylococcus aureus transcriptome meta-database (SATMD); Transcript regulatory network; Transcriptome.
Figures

References
-
- Francis JS, Doherty MC, Lopatin U, Johnston CP, Sinha G, Ross T, Cai M, Hansel NN, Perl T, Ticehurst JR, Carroll K, Thomas DL, Nuermberger E, Bartlett JG. Severe community-onset pneumonia in healthy adults caused by methicillin-resistant Staphylococcus aureus carrying the Panton-Valentine leukocidin genes. Clinical Infectious Diseases. 2005;40:100–107. doi: 10.1086/427148. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

