Down-regulation of Fusarium oxysporum endogenous genes by Host-Delivered RNA interference enhances disease resistance
- PMID: 25654075
- PMCID: PMC4299518
- DOI: 10.3389/fchem.2015.00001
Down-regulation of Fusarium oxysporum endogenous genes by Host-Delivered RNA interference enhances disease resistance
Abstract
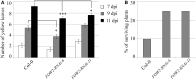
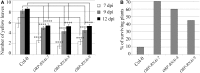
Fusarium oxysporum is a devastating pathogen causing extensive yield losses in a variety of crops and development of sustainable, environmentally friendly methods to improve crop resistance is crucial. We have used Host-Delivered RNA interference (HD-RNAi) technology to partially silence three different genes (FOW2, FRP1, and OPR) in the hemi-biotrophic fungus F. oxysporum f. sp. conglutinans. Expression of double stranded RNA (dsRNA) molecules targeting fungal pathogen genes was achieved in a number of transgenic Arabidopsis lines. F. oxysporum infecting the transgenic lines displayed substantially reduced mRNA levels on all three targeted genes, with an average of 75, 83, and 72% reduction for FOW2, FRP1, and OPR, respectively. The silencing of pathogen genes had a clear positive effect on the ability of the transgenic lines to fight infection. All transgenic lines displayed enhanced resistance to F. oxysporum with delayed disease symptom development, especially FRP1 and OPR lines. Survival rates after fungal infection were higher in the transgenic lines compared to control wild type plants which consistently showed survival rates of 10%, with FOW2 lines showing 25% survival; FRP1 lines 30-50% survival and OPR between 45 and 70% survival. The down-regulation effect was specific for the targeted genes without unintended effects in related genes. In addition to producing resistant crops, HD-RNAi can provide a useful tool to rapidly screen candidate fungal pathogenicity genes without the need to produce fungal knockout mutants.
Keywords: Fusarium oxysporum; Host-Delivered RNAi; disease control; disease resistance; host-induced gene silencing; plant fungal pathogens.
Figures



Similar articles
-
Host-Induced Silencing of Pathogenicity Genes Enhances Resistance to Fusarium oxysporum Wilt in Tomato.Mol Biotechnol. 2017 Aug;59(8):343-352. doi: 10.1007/s12033-017-0022-y. Mol Biotechnol. 2017. PMID: 28674943
-
Fow2, a Zn(II)2Cys6-type transcription regulator, controls plant infection of the vascular wilt fungus Fusarium oxysporum.Mol Microbiol. 2007 Feb;63(3):737-53. doi: 10.1111/j.1365-2958.2006.05554.x. Mol Microbiol. 2007. PMID: 17302801
-
RNAi-mediated silencing of PEX6 and GAS1 genes of Fusarium oxysporum f. sp. lycopersici confers resistance against Fusarium wilt in tomato.3 Biotech. 2021 Oct;11(10):443. doi: 10.1007/s13205-021-02973-8. Epub 2021 Sep 21. 3 Biotech. 2021. PMID: 34631344 Free PMC article.
-
RNA Interference (RNAi) as a Potential Tool for Control of Mycotoxin Contamination in Crop Plants: Concepts and Considerations.Front Plant Sci. 2017 Feb 14;8:200. doi: 10.3389/fpls.2017.00200. eCollection 2017. Front Plant Sci. 2017. PMID: 28261252 Free PMC article. Review.
-
Mycovirus-encoded suppressors of RNA silencing: Possible allies or enemies in the use of RNAi to control fungal disease in crops.Front Fungal Biol. 2022 Oct 10;3:965781. doi: 10.3389/ffunb.2022.965781. eCollection 2022. Front Fungal Biol. 2022. PMID: 37746227 Free PMC article. Review.
Cited by
-
RNA Interference Strategies for Future Management of Plant Pathogenic Fungi: Prospects and Challenges.Plants (Basel). 2021 Mar 29;10(4):650. doi: 10.3390/plants10040650. Plants (Basel). 2021. PMID: 33805521 Free PMC article.
-
Concepts and considerations for enhancing RNAi efficiency in phytopathogenic fungi for RNAi-based crop protection using nanocarrier-mediated dsRNA delivery systems.Front Fungal Biol. 2022 Sep 8;3:977502. doi: 10.3389/ffunb.2022.977502. eCollection 2022. Front Fungal Biol. 2022. PMID: 37746174 Free PMC article. Review.
-
Sclerotinia sclerotiorum Thioredoxin1 (SsTrx1) is required for pathogenicity and oxidative stress tolerance.Mol Plant Pathol. 2021 Nov;22(11):1413-1426. doi: 10.1111/mpp.13127. Epub 2021 Aug 30. Mol Plant Pathol. 2021. PMID: 34459563 Free PMC article.
-
Host induced gene silencing of Magnaporthe oryzae by targeting pathogenicity and development genes to control rice blast disease.Front Plant Sci. 2022 Aug 11;13:959641. doi: 10.3389/fpls.2022.959641. eCollection 2022. Front Plant Sci. 2022. PMID: 36035704 Free PMC article.
-
RNA-Spray-Mediated Silencing of Fusarium graminearum AGO and DCL Genes Improve Barley Disease Resistance.Front Plant Sci. 2020 Apr 29;11:476. doi: 10.3389/fpls.2020.00476. eCollection 2020. Front Plant Sci. 2020. PMID: 32411160 Free PMC article.
References
-
- Aly R., Cholakh H., Joel D. M., Leibman D., Steinitz B., Zelcer A., et al. . (2009). Gene silencing of mannose 6-phosphate reductase in the parasitic weed Orobanche aegyptiaca through the production of homologous dsRNA sequences in the host plant. Plant Biotech. J. 7, 487–498. 10.1111/j.1467-7652.2009.00418.x - DOI - PubMed
-
- Anderson J. P., Badruzsaufari E., Schenk P. M., Manners J. M., Desmond O. J., Ehlert C., et al. . (2004). Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 16, 3460–3479. 10.1105/tpc.104.025833 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

