Replacing uridine with 2-thiouridine enhances the rate and fidelity of nonenzymatic RNA primer extension
- PMID: 25654265
- PMCID: PMC4985000
- DOI: 10.1021/jacs.5b00445
Replacing uridine with 2-thiouridine enhances the rate and fidelity of nonenzymatic RNA primer extension
Abstract
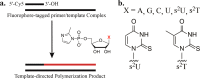
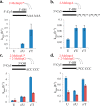
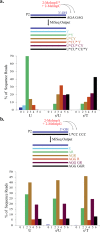
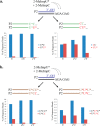
The nonenzymatic replication of RNA oligonucleotides is thought to have played a key role in the origin of life prior to the evolution of ribozyme-catalyzed RNA replication. Although the copying of oligo-C templates by 2-methylimidazole-activated G monomers can be quite efficient, the copying of mixed sequence templates, especially those containing A and U, is particularly slow and error-prone. The greater thermodynamic stability of the 2-thio-U(s(2)U):A base pair, relative to the canonical U:A base pair, suggests that replacing U with s(2)U might enhance the rate and fidelity of the nonenzymatic copying of RNA templates. Here we report that this single atom substitution in the activated monomer improves both the kinetics and the fidelity of nonenzymatic primer extension on mixed-sequence RNA templates. In addition, the mean lengths of primer extension products obtained with s(2)U is greater than those obtained with U, augmenting the potential for nonenzymatic replication of heritable function-rich sequences. We suggest that noncanonical nucleotides such as s(2)U may have played a role during the infancy of the RNA world by facilitating the nonenzymatic replication of genomic RNA oligonucleotides.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

Similar articles
-
Unusual Base Pair between Two 2-Thiouridines and Its Implication for Nonenzymatic RNA Copying.J Am Chem Soc. 2024 Feb 14;146(6):3861-3871. doi: 10.1021/jacs.3c11158. Epub 2024 Jan 31. J Am Chem Soc. 2024. PMID: 38293747 Free PMC article.
-
Nonenzymatic RNA copying with a potentially primordial genetic alphabet.Proc Natl Acad Sci U S A. 2025 May 27;122(21):e2505720122. doi: 10.1073/pnas.2505720122. Epub 2025 May 21. Proc Natl Acad Sci U S A. 2025. PMID: 40397670 Free PMC article.
-
Thiolated uridine substrates and templates improve the rate and fidelity of ribozyme-catalyzed RNA copying.Chem Commun (Camb). 2016 May 5;52(39):6529-32. doi: 10.1039/c6cc02692c. Chem Commun (Camb). 2016. PMID: 27109314
-
The Emergence of RNA from the Heterogeneous Products of Prebiotic Nucleotide Synthesis.J Am Chem Soc. 2021 Mar 10;143(9):3267-3279. doi: 10.1021/jacs.0c12955. Epub 2021 Feb 26. J Am Chem Soc. 2021. PMID: 33636080 Review.
-
The Mechanism of Nonenzymatic Template Copying with Imidazole-Activated Nucleotides.Angew Chem Int Ed Engl. 2019 Aug 5;58(32):10812-10819. doi: 10.1002/anie.201902050. Epub 2019 Jul 4. Angew Chem Int Ed Engl. 2019. PMID: 30908802 Review.
Cited by
-
Inosine in Biology and Disease.Genes (Basel). 2021 Apr 19;12(4):600. doi: 10.3390/genes12040600. Genes (Basel). 2021. PMID: 33921764 Free PMC article. Review.
-
Inosine, but none of the 8-oxo-purines, is a plausible component of a primordial version of RNA.Proc Natl Acad Sci U S A. 2018 Dec 26;115(52):13318-13323. doi: 10.1073/pnas.1814367115. Epub 2018 Dec 3. Proc Natl Acad Sci U S A. 2018. PMID: 30509978 Free PMC article.
-
Taming Prebiotic Chemistry: The Role of Heterogeneous and Interfacial Catalysis in the Emergence of a Prebiotic Catalytic/Information Polymer System.Life (Basel). 2016 Nov 4;6(4):40. doi: 10.3390/life6040040. Life (Basel). 2016. PMID: 27827919 Free PMC article. Review.
-
Overcoming nucleotide bias in the nonenzymatic copying of RNA templates.Nucleic Acids Res. 2024 Dec 11;52(22):13515-13529. doi: 10.1093/nar/gkae982. Nucleic Acids Res. 2024. PMID: 39530216 Free PMC article.
-
Kinetic explanations for the sequence biases observed in the nonenzymatic copying of RNA templates.Nucleic Acids Res. 2022 Jan 11;50(1):35-45. doi: 10.1093/nar/gkab1202. Nucleic Acids Res. 2022. PMID: 34893864 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

