Essential role for autophagy in life span extension
- PMID: 25654554
- PMCID: PMC4382258
- DOI: 10.1172/JCI73946
Essential role for autophagy in life span extension
Abstract
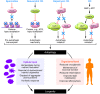
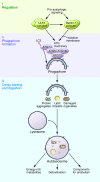
Life and health span can be prolonged by calorie limitation or by pharmacologic agents that mimic the effects of caloric restriction. Both starvation and the genetic inactivation of nutrient signaling converge on the induction of autophagy, a cytoplasmic recycling process that counteracts the age-associated accumulation of damaged organelles and proteins as it improves the metabolic fitness of cells. Here we review experimental findings indicating that inhibition of the major nutrient and growth-related signaling pathways as well as the upregulation of anti-aging pathways mediate life span extension via the induction of autophagy. Furthermore, we discuss mounting evidence suggesting that autophagy is not only necessary but, at least in some cases, also sufficient for increasing longevity.
Figures

Similar articles
-
Autophagy and aging.Cell. 2011 Sep 2;146(5):682-95. doi: 10.1016/j.cell.2011.07.030. Cell. 2011. PMID: 21884931 Review.
-
Towards an understanding of the anti-aging mechanism of caloric restriction.Curr Aging Sci. 2008 Mar;1(1):4-9. doi: 10.2174/1874609810801010004. Curr Aging Sci. 2008. PMID: 20021367
-
Longevity pathways converge on autophagy genes to regulate life span in Caenorhabditis elegans.Autophagy. 2008 Apr;4(3):330-8. doi: 10.4161/auto.5618. Epub 2008 Jan 21. Autophagy. 2008. PMID: 18219227
-
The role of autophagy in aging: its essential part in the anti-aging mechanism of caloric restriction.Ann N Y Acad Sci. 2007 Oct;1114:69-78. doi: 10.1196/annals.1396.020. Epub 2007 Oct 12. Ann N Y Acad Sci. 2007. PMID: 17934054 Review.
-
Autophagy is required for dietary restriction-mediated life span extension in C. elegans.Autophagy. 2007 Nov-Dec;3(6):597-9. doi: 10.4161/auto.4989. Epub 2007 Sep 6. Autophagy. 2007. PMID: 17912023
Cited by
-
Natural Products in the Promotion of Healthspan and Longevity.Clin Pharmacol Transl Med. 2019;3(1):149-151. doi: 10.31700/2572-7656.000123. Epub 2019 Jul 16. Clin Pharmacol Transl Med. 2019. PMID: 31363716 Free PMC article. No abstract available.
-
Lifespan-increasing drug nordihydroguaiaretic acid inhibits p300 and activates autophagy.NPJ Aging Mech Dis. 2019 Oct 2;5:7. doi: 10.1038/s41514-019-0037-7. eCollection 2019. NPJ Aging Mech Dis. 2019. PMID: 31602311 Free PMC article.
-
Influence of the Mediterranean Diet on Healthy Aging.Int J Mol Sci. 2023 Feb 24;24(5):4491. doi: 10.3390/ijms24054491. Int J Mol Sci. 2023. PMID: 36901921 Free PMC article. Review.
-
Branching Off: New Insight Into Lysosomes as Tubular Organelles.Front Cell Dev Biol. 2022 May 11;10:863922. doi: 10.3389/fcell.2022.863922. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35646899 Free PMC article. Review.
-
Calorie Restriction Rescues Mitochondrial Dysfunction in Adck2-Deficient Skeletal Muscle.Front Physiol. 2022 Jul 14;13:898792. doi: 10.3389/fphys.2022.898792. eCollection 2022. Front Physiol. 2022. PMID: 35936917 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

