Characterizing sialic acid variants at the glycopeptide level
- PMID: 25654559
- PMCID: PMC4367445
- DOI: 10.1021/ac504725r
Characterizing sialic acid variants at the glycopeptide level
Abstract
Beam-type collision-induced dissociation (CID) data of intact glycopeptides isolated from mouse liver tissue are presented to illustrate characteristic fragmentation of differentially sialylated glycopeptides. Eight glycoforms of an O-linked glycopeptide from Nucleobindin-1 are distinguished on the basis of the precursor masses and characteristic oxonium ions. We report that all sialic acid variants are prone to neutral loss from the charge reduced species in electron-transfer dissociation (ETD) fragmentation. We show how changes in sialic acid composition affect reverse phase chromatographic retention times: sialic acid addition increases glycopeptide retention times significantly; replacing the N-acetylneuraminic acid with the N-glycolyl variant leads to slightly reduced retention times, while O-acetylated sialic acid-containing glycoforms are retained longer. We then demonstrate how MS-Filter in Protein Prospector can use these diagnostic oxonium ions to find glycopeptides, by showing that a wealth of different glycopeptides can be found in a published phosphopeptide data set.
Figures
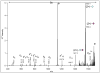

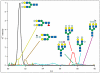
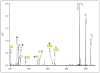
Similar articles
-
Sialic acid capture-and-release and LC-MS(n) analysis of glycopeptides.Methods Mol Biol. 2013;951:79-100. doi: 10.1007/978-1-62703-146-2_7. Methods Mol Biol. 2013. PMID: 23296526
-
Improved online LC-MS/MS identification of O-glycosites by EThcD fragmentation, chemoenzymatic reaction, and SPE enrichment.Glycoconj J. 2021 Apr;38(2):145-156. doi: 10.1007/s10719-020-09952-w. Epub 2020 Oct 17. Glycoconj J. 2021. PMID: 33068214 Free PMC article.
-
Glycan side reaction may compromise ETD-based glycopeptide identification.J Am Soc Mass Spectrom. 2014 Jun;25(6):977-87. doi: 10.1007/s13361-014-0852-9. Epub 2014 Mar 25. J Am Soc Mass Spectrom. 2014. PMID: 24664807 Free PMC article.
-
[Recent advances in glycopeptide enrichment and mass spectrometry data interpretation approaches for glycoproteomics analyses].Se Pu. 2021 Oct;39(10):1045-1054. doi: 10.3724/SP.J.1123.2021.06011. Se Pu. 2021. PMID: 34505426 Free PMC article. Review. Chinese.
-
Liquid chromatography-tandem mass spectrometry-based fragmentation analysis of glycopeptides.Glycoconj J. 2016 Jun;33(3):261-72. doi: 10.1007/s10719-016-9649-3. Epub 2016 Jan 18. Glycoconj J. 2016. PMID: 26780731 Review.
Cited by
-
O-glycosylation sites identified from mucin core-1 type glycopeptides from human serum.Glycoconj J. 2016 Jun;33(3):435-45. doi: 10.1007/s10719-015-9630-6. Epub 2016 Jan 5. Glycoconj J. 2016. PMID: 26729242 Free PMC article.
-
Discovery and Characterization of Unusual O-Linked Glycosylation of IgG4 Antibody Using LC-MS.Rapid Commun Mass Spectrom. 2025 Mar;39(5):e9969. doi: 10.1002/rcm.9969. Rapid Commun Mass Spectrom. 2025. PMID: 39663547 Free PMC article.
-
Tissue-Specific Glycosylation at the Glycopeptide Level.Mol Cell Proteomics. 2015 Aug;14(8):2103-10. doi: 10.1074/mcp.M115.050393. Epub 2015 May 20. Mol Cell Proteomics. 2015. PMID: 25995273 Free PMC article.
-
Maturing Glycoproteomics Technologies Provide Unique Structural Insights into the N-glycoproteome and Its Regulation in Health and Disease.Mol Cell Proteomics. 2016 Jun;15(6):1773-90. doi: 10.1074/mcp.O115.057638. Epub 2016 Feb 29. Mol Cell Proteomics. 2016. PMID: 26929216 Free PMC article. Review.
-
Proteomic analysis reveals O-GlcNAc modification on proteins with key regulatory functions in Arabidopsis.Proc Natl Acad Sci U S A. 2017 Feb 21;114(8):E1536-E1543. doi: 10.1073/pnas.1610452114. Epub 2017 Feb 2. Proc Natl Acad Sci U S A. 2017. PMID: 28154133 Free PMC article.
References
-
- Apweiler R, Hermjakob H, Sharon N. Biochim Biophys Acta. 1999;1473:4–8. - PubMed
-
- Jacob GS, Welply JK, Scudder PR, Kirmaier C, Abbas SZ, Howard SC, Keene JL, Schmuke JJ, Broschat K, Steininger C. Adv Exp Med Biol. 1995;376:283–290. - PubMed
-
- Woronowicz A, Amith SR, De Vusser K, Laroy W, Contreras R, Basta S, Szewczuk MR. Glycobiology. 2007;17:10–24. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

