On the biased nucleotide composition of the human coronavirus RNA genome
- PMID: 25656063
- PMCID: PMC7114406
- DOI: 10.1016/j.virusres.2014.11.031
On the biased nucleotide composition of the human coronavirus RNA genome
Abstract
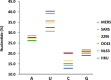
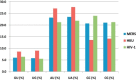
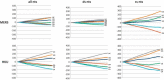
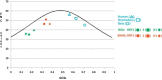
We investigated the nucleotide composition of the RNA genome of the six human coronaviruses. Some general coronavirus characteristics were apparent (e.g. high U, low C count), but we also detected species-specific signatures. Most strikingly, the high U and low C proportions are quite variable and act like communicating vessels, C goes down when U goes up and vice versa. U ranges among virus isolates from 30.7% to 40.3%, and C makes the opposite movement from 20.0% to 12.9%, respectively. The nucleotide biases are more pronounced in the unpaired regions of the structured RNA genome, which may suggest a certain biological function for these distinctive sequence signatures. Coronaviruses have an atypical codon usage that has been linked to mutational events operating on the viral RNA genome on an evolutionary time scale. We suggest that the atypical nucleotide bias may serve a distinct biological function and that it is the direct cause of the characteristic codon usage in these viruses. The relevance for evolution of the novel human pathogens MERS and SARS is discussed.
Keywords: Coronavirus; MERS; Nucleotide signature; RNA genome; SARS.
Copyright © 2015 Elsevier B.V. All rights reserved.
Figures
References
-
- Bennetzen J.L., Hall B.D. Codon selection in yeast. J. Biol. Chem. 1982;257:3026–3031. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

