Signatures of adaptation to plant parasitism in nematode genomes
- PMID: 25656361
- PMCID: PMC4413825
- DOI: 10.1017/S0031182013002163
Signatures of adaptation to plant parasitism in nematode genomes
Abstract
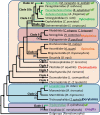
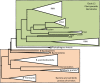
Plant-parasitic nematodes cause considerable damage to global agriculture. The ability to parasitize plants is a derived character that appears to have independently emerged several times in the phylum Nematoda. Morphological convergence to feeding style has been observed, but whether this is emergent from molecular convergence is less obvious. To address this, we assess whether genomic signatures can be associated with plant parasitism by nematodes. In this review, we report genomic features and characteristics that appear to be common in plant-parasitic nematodes while absent or rare in animal parasites, predators or free-living species. Candidate horizontal acquisitions of parasitism genes have systematically been found in all plant-parasitic species investigated at the sequence level. Presence of peptides that mimic plant hormones also appears to be a trait of plant-parasitic species. Annotations of the few genomes of plant-parasitic nematodes available to date have revealed a set of apparently species-specific genes on every occasion. Effector genes, important for parasitism are frequently found among those species-specific genes, indicating poor overlap. Overall, nematodes appear to have developed convergent genomic solutions to adapt to plant parasitism.
Keywords: convergence.
Figures

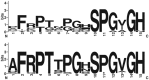

References
-
- Abad P., Gouzy J., Aury J.-M., Castagnone-Sereno P., Danchin E. G. J., Deleury E., Perfus-Barbeoch L., Anthouard V., Artiguenave F., Blok V. C., Caillaud M.-C., Coutinho P. M., Dasilva C., Luca F. D., Deau F., Esquibet M., Flutre T., Goldstone J. V., Hamamouch N., Hewezi T., Jaillon O., Jubin C., Leonetti P., Magliano M., Maier T. R., Markov G. V., McVeigh P., Pesole G., Poulain J., Robinson-Rechavi M., Sallet E., Ségurens B., Steinbach D., Tytgat T., Ugarte E., van Ghelder C., Veronico P., Baum T. J., Blaxter M., Bleve-Zacheo T., Davis E. L., Ewbank J. J., Favery B., Grenier E., Henrissat B., Jones J. T., Laudet V., Maule A. G., Quesneville H., Rosso M.-N., Schiex T., Smant G., Weissenbach J. and Wincker P. (2008). Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nature Biotechnology 26, 909–915. doi: 10.1038/nbt.1482. - DOI - PubMed
-
- Baldwin J. G., Nadler S. A. and Adams B. J. (2004). Evolution of plant parasitism among nematodes. Annual Review of Phytopathology 42, 83–105. - PubMed
-
- Bird D. M. (1996). Manipulation of host gene expression by root-knot nematodes. Journal of Parasitology 82, 881–888. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

