Application of clinical text data for phenome-wide association studies (PheWASs)
- PMID: 25657332
- PMCID: PMC4481696
- DOI: 10.1093/bioinformatics/btv076
Application of clinical text data for phenome-wide association studies (PheWASs)
Abstract
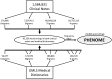
Motivation: Genome-wide association studies (GWASs) are effective for describing genetic complexities of common diseases. Phenome-wide association studies (PheWASs) offer an alternative and complementary approach to GWAS using data embedded in the electronic health record (EHR) to define the phenome. International Classification of Disease version 9 (ICD9) codes are used frequently to define the phenome, but using ICD9 codes alone misses other clinically relevant information from the EHR that can be used for PheWAS analyses and discovery.
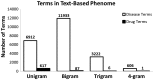
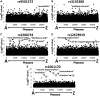
Results: As an alternative to ICD9 coding, a text-based phenome was defined by 23 384 clinically relevant terms extracted from Marshfield Clinic's EHR. Five single nucleotide polymorphisms (SNPs) with known phenotypic associations were genotyped in 4235 individuals and associated across the text-based phenome. All five SNPs genotyped were associated with expected terms (P<0.02), most at or near the top of their respective PheWAS ranking. Raw association results indicate that text data performed equivalently to ICD9 coding and demonstrate the utility of information beyond ICD9 coding for application in PheWAS.
© The Author 2015. Published by Oxford University Press. All rights reserved. For Permissions, please e-mail: journals.permissions@oup.com.
Figures

References
Publication types
MeSH terms
Grants and funding
- K22 LM011938/LM/NLM NIH HHS/United States
- UL1 TR000427/TR/NCATS NIH HHS/United States
- 1U01HG006389/HG/NHGRI NIH HHS/United States
- T15 LM007359/LM/NLM NIH HHS/United States
- R01GM097618/GM/NIGMS NIH HHS/United States
- U01 HG006389/HG/NHGRI NIH HHS/United States
- 1UL1RR025011/RR/NCRR NIH HHS/United States
- 9U54TR000021/TR/NCATS NIH HHS/United States
- 1K22LM011938/LM/NLM NIH HHS/United States
- R01 GM097618/GM/NIGMS NIH HHS/United States
- UL1 RR025011/RR/NCRR NIH HHS/United States
- 5T15LM007359,/LM/NLM NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

