Curve interpolation model for visualising disjointed neural elements
- PMID: 25657704
- PMCID: PMC4308766
- DOI: 10.3969/j.issn.1673-5374.2012.21.006
Curve interpolation model for visualising disjointed neural elements
Abstract
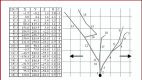
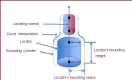
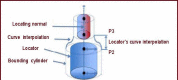
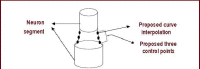
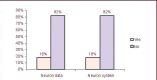
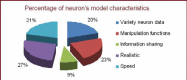
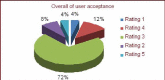
Neuron cell are built from a myriad of axon and dendrite structures. It transmits electrochemical signals between the brain and the nervous system. Three-dimensional visualization of neuron structure could help to facilitate deeper understanding of neuron and its models. An accurate neuron model could aid understanding of brain's functionalities, diagnosis and knowledge of entire nervous system. Existing neuron models have been found to be defective in the aspect of realism. Whereas in the actual biological neuron, there is continuous growth as the soma extending to the axon and the dendrite; but, the current neuron visualization models present it as disjointed segments that has greatly mediated effective realism. In this research, a new reconstruction model comprising of the Bounding Cylinder, Curve Interpolation and Gouraud Shading is proposed to visualize neuron model in order to improve realism. The reconstructed model is used to design algorithms for generating neuron branching from neuron SWC data. The Bounding Cylinder and Curve Interpolation methods are used to improve the connected segments of the neuron model using a series of cascaded cylinders along the neuron's connection path. Three control points are proposed between two adjacent neuron segments. Finally, the model is rendered with Gouraud Shading for smoothening of the model surface. This produce a near-perfection model of the natural neurons with attended realism. The model is validated by a group of bioinformatics analysts' responses to a predefined survey. The result shows about 82% acceptance and satisfaction rate.
Keywords: Gouraud shading; bounding cylinder; curve interpolation; neural regeneration; reconstruction model.
Conflict of interest statement
Figures








Similar articles
-
Automated 3D Soma Segmentation with Morphological Surface Evolution for Neuron Reconstruction.Neuroinformatics. 2018 Apr;16(2):153-166. doi: 10.1007/s12021-017-9353-x. Neuroinformatics. 2018. PMID: 29344781
-
Ultrastructural basis of strong unitary inhibition in a binaural neuron.J Physiol. 2018 Oct;596(20):4969-4982. doi: 10.1113/JP276015. Epub 2018 Sep 2. J Physiol. 2018. PMID: 30054922 Free PMC article.
-
Faster shading by equal angle interpolation of vectors.IEEE Trans Vis Comput Graph. 2004 Mar-Apr;10(2):217-23. doi: 10.1109/TVCG.2004.1260773. IEEE Trans Vis Comput Graph. 2004. PMID: 15384646
-
The Dorsal Column Lesion Model of Spinal Cord Injury and Its Use in Deciphering the Neuron-Intrinsic Injury Response.Dev Neurobiol. 2018 Oct;78(10):926-951. doi: 10.1002/dneu.22601. Epub 2018 May 11. Dev Neurobiol. 2018. PMID: 29717546 Free PMC article. Review.
-
Mechanisms and molecules in motor neuron specification and axon pathfinding.Bioessays. 2001 Jul;23(7):582-95. doi: 10.1002/bies.1084. Bioessays. 2001. PMID: 11462212 Review.
References
-
- Johnson CR, MacLeod R, Parker SG, et al. Biomedical computing and visualization software environment. Comm ACM. 2004;47(11):65–71.
-
- Rehman A, Saba T. Off-line cursive script recognition: current advances, comparisons and remaining problems Artif Intell Rev. 2012;37(4):261–268.
-
- Haron H, Rehman A, Adi DIS, et al. Parameterization method on B-spline curve. Math Probl Eng. 2012 doi:10.1155/2012/640472.
-
- Ascoli GA, Scorcioni R. Neuron and Network Modeling. In: Zaborszky L, Wouterlood FG, Lanciego JL, editors. Neuroanatomical Tract-Tracing 3: Molecules, Neurons and Systems. Berlin: Springer Publisher; 2006.
LinkOut - more resources
Full Text Sources
