Glioma-associated microglia/macrophages display an expression profile different from M1 and M2 polarization and highly express Gpnmb and Spp1
- PMID: 25658639
- PMCID: PMC4320099
- DOI: 10.1371/journal.pone.0116644
Glioma-associated microglia/macrophages display an expression profile different from M1 and M2 polarization and highly express Gpnmb and Spp1
Abstract
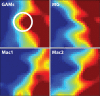
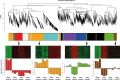
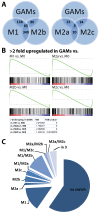
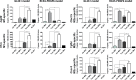
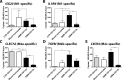
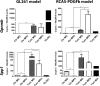
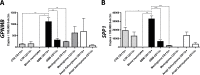
Malignant glioma belong to the most aggressive neoplasms in humans with no successful treatment available. Patients suffering from glioblastoma multiforme (GBM), the highest-grade glioma, have an average survival time of only around one year after diagnosis. Both microglia and peripheral macrophages/monocytes accumulate within and around glioma, but fail to exert effective anti-tumor activity and even support tumor growth. Here we use microarray analysis to compare the expression profiles of glioma-associated microglia/macrophages and naive control cells. Samples were generated from CD11b+ MACS-isolated cells from naïve and GL261-implanted C57BL/6 mouse brains. Around 1000 genes were more than 2-fold up- or downregulated in glioma-associated microglia/macrophages when compared to control cells. A comparison with published data sets of M1, M2a,b,c-polarized macrophages revealed a gene expression pattern that has only partial overlap with any of the M1 or M2 gene expression patterns. Samples for the qRT-PCR validation of selected M1 and M2a,b,c-specific genes were generated from two different glioma mouse models and isolated by flow cytometry to distinguish between resident microglia and invading macrophages. We confirmed in both models the unique glioma-associated microglia/macrophage phenotype including a mixture of M1 and M2a,b,c-specific genes. To validate the expression of these genes in human we MACS-isolated CD11b+ microglia/macrophages from GBM, lower grade brain tumors and control specimens. Apart from the M1/M2 gene analysis, we demonstrate that the expression of Gpnmb and Spp1 is highly upregulated in both murine and human glioma-associated microglia/macrophages. High expression of these genes has been associated with poor prognosis in human GBM, as indicated by patient survival data linked to gene expression data. We also show that microglia/macrophages are the predominant source of these transcripts in murine and human GBM. Our findings provide new potential targets for future anti-glioma therapy.
Conflict of interest statement
Figures


References
-
- Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, et al. (2006) Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer cell 9: 157–173. - PubMed
-
- Sottoriva A, Spiteri I, Piccirillo SG, Touloumis A, Collins VP, et al. (2013) Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proceedings of the National Academy of Sciences of the United States of America 110: 4009–4014. 10.1073/pnas.1219747110 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

