Use of 16S rRNA gene for identification of a broad range of clinically relevant bacterial pathogens
- PMID: 25658760
- PMCID: PMC4319838
- DOI: 10.1371/journal.pone.0117617
Use of 16S rRNA gene for identification of a broad range of clinically relevant bacterial pathogens
Abstract
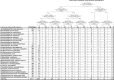
According to World Health Organization statistics of 2011, infectious diseases remain in the top five causes of mortality worldwide. However, despite sophisticated research tools for microbial detection, rapid and accurate molecular diagnostics for identification of infection in humans have not been extensively adopted. Time-consuming culture-based methods remain to the forefront of clinical microbial detection. The 16S rRNA gene, a molecular marker for identification of bacterial species, is ubiquitous to members of this domain and, thanks to ever-expanding databases of sequence information, a useful tool for bacterial identification. In this study, we assembled an extensive repository of clinical isolates (n = 617), representing 30 medically important pathogenic species and originally identified using traditional culture-based or non-16S molecular methods. This strain repository was used to systematically evaluate the ability of 16S rRNA for species level identification. To enable the most accurate species level classification based on the paucity of sequence data accumulated in public databases, we built a Naïve Bayes classifier representing a diverse set of high-quality sequences from medically important bacterial organisms. We show that for species identification, a model-based approach is superior to an alignment based method. Overall, between 16S gene based and clinical identities, our study shows a genus-level concordance rate of 96% and a species-level concordance rate of 87.5%. We point to multiple cases of probable clinical misidentification with traditional culture based identification across a wide range of gram-negative rods and gram-positive cocci as well as common gram-negative cocci.
Conflict of interest statement
Figures




References
-
- Martin GS, Mannino DM, Eaton S, Moss M (2003) The epidemiology of sepsis in the United States from 1979 through 2000. The New England journal of medicine 348: 1546–1554. - PubMed
-
- Iregui M, Ward S, Sherman G, Fraser VJ, Kollef MH (2002) Clinical importance of delays in the initiation of appropriate antibiotic treatment for ventilator-associated pneumonia. Chest 122: 262–268. - PubMed
-
- Larson E (2007) Community factors in the development of antibiotic resistance. Annu Rev Public Health 28: 435–447. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

