The major autoantibody epitope on factor H in atypical hemolytic uremic syndrome is structurally different from its homologous site in factor H-related protein 1, supporting a novel model for induction of autoimmunity in this disease
- PMID: 25659429
- PMCID: PMC4392255
- DOI: 10.1074/jbc.M114.630871
The major autoantibody epitope on factor H in atypical hemolytic uremic syndrome is structurally different from its homologous site in factor H-related protein 1, supporting a novel model for induction of autoimmunity in this disease
Abstract
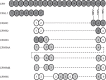
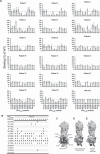
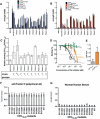
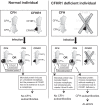
Atypical hemolytic uremic syndrome (aHUS) is characterized by complement attack against host cells due to mutations in complement proteins or autoantibodies against complement factor H (CFH). It is unknown why nearly all patients with autoimmune aHUS lack CFHR1 (CFH-related protein-1). These patients have autoantibodies against CFH domains 19 and 20 (CFH19-20), which are nearly identical to CFHR1 domains 4 and 5 (CFHR14-5). Here, binding site mapping of autoantibodies from 17 patients using mutant CFH19-20 constructs revealed an autoantibody epitope cluster within a loop on domain 20, next to the two buried residues that are different in CFH19-20 and CFHR14-5. The crystal structure of CFHR14-5 revealed a difference in conformation of the autoantigenic loop in the C-terminal domains of CFH and CFHR1, explaining the variation in binding of autoantibodies from some aHUS patients to CFH19-20 and CFHR14-5. The autoantigenic loop on CFH seems to be generally flexible, as its conformation in previously published structures of CFH19-20 bound to the microbial protein OspE and a sialic acid glycan is somewhat altered. Cumulatively, our data suggest that association of CFHR1 deficiency with autoimmune aHUS could be due to the structural difference between CFHR1 and the autoantigenic CFH epitope, suggesting a novel explanation for CFHR1 deficiency in the pathogenesis of autoimmune aHUS.
Keywords: Atypical Hemolytic Uremic Syndrome; Autoimmune Disease; Autoimmunity; Complement Regulation; Immunoglobulin G (IgG); Immunology; Structure-Function Study; Thrombotic Microangiopathy; X-ray Crystallography.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


References
-
- Noris M., Remuzzi G. (2009) Atypical hemolytic-uremic syndrome. N. Engl. J. Med. 361, 1676–1687 - PubMed
-
- Sánchez-Corral P., Melgosa M. (2010) Advances in understanding the aetiology of atypical haemolytic uraemic Syndrome. Br. J. Haematol 150, 529–542 - PubMed
-
- Zipfel P. F., Edey M., Heinen S., Józsi M., Richter H., Misselwitz J., Hoppe B., Routledge D., Strain L., Hughes A. E., Goodship J. A., Licht C., Goodship T. H., Skerka C. (2007) Deletion of complement factor H-related genes CFHR1 and CFHR3 is associated with atypical hemolytic uremic syndrome. PLoS Genet 3, e41. - PMC - PubMed
-
- Venables J. P., Strain L., Routledge D., Bourn D., Powell H. M., Warwicker P., Diaz-Torres M. L., Sampson A., Mead P., Webb M., Pirson Y., Jackson M. S., Hughes A., Wood K. M., Goodship J. A., Goodship T. H. (2006) Atypical haemolytic uraemic syndrome associated with a hybrid complement gene. PLoS Med. 3, e431. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

