Rph1/KDM4 mediates nutrient-limitation signaling that leads to the transcriptional induction of autophagy
- PMID: 25660547
- PMCID: PMC4348152
- DOI: 10.1016/j.cub.2014.12.049
Rph1/KDM4 mediates nutrient-limitation signaling that leads to the transcriptional induction of autophagy
Abstract
Background: Autophagy is a conserved process mediating vacuolar degradation and recycling. Autophagy is highly upregulated upon various stresses and is essential for cell survival in deleterious conditions. Autophagy defects are associated with severe pathologies, whereas unchecked autophagy activity causes cell death. Therefore, to support proper cellular homeostasis, the induction and amplitude of autophagy activity have to be finely regulated. Transcriptional control is a critical, yet largely unexplored, aspect of autophagy regulation. In particular, little is known about the signaling pathways modulating the expression of autophagy-related genes, and only a few transcriptional regulators have been identified as contributing in the control of this process.
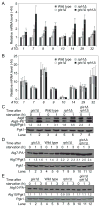
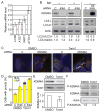
Results: We identified Rph1 as a negative regulator of the transcription of several ATG genes and a repressor of autophagy induction. Rph1 is a histone demethylase protein, but it regulates autophagy independently of its demethylase activity. Rim15 mediates the phosphorylation of Rph1 upon nitrogen starvation, which causes an inhibition of its function. Preventing Rph1 phosphorylation or overexpressing the protein causes a severe block in autophagy induction. A similar function of Rph1/KDM4 is seen in mammalian cells, indicating that this process is highly conserved.
Conclusion: Rph1 maintains autophagy at a low level in nutrient-rich conditions; upon nutrient limitation, the inhibition of its activity is a prerequisite to the induction of ATG gene transcription and autophagy.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures





Comment in
-
Identifying Autophagy-Associated Proteins and Chemicals with a Random Walk-Based Method within Heterogeneous Interaction Network.Front Biosci (Landmark Ed). 2024 Jan 17;29(1):21. doi: 10.31083/j.fbl2901021. Front Biosci (Landmark Ed). 2024. PMID: 38287832
References
-
- Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. - PubMed
-
- Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. - PubMed
-
- Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

