FF483-484 motif of human Polη mediates its interaction with the POLD2 subunit of Polδ and contributes to DNA damage tolerance
- PMID: 25662213
- PMCID: PMC4344513
- DOI: 10.1093/nar/gkv076
FF483-484 motif of human Polη mediates its interaction with the POLD2 subunit of Polδ and contributes to DNA damage tolerance
Abstract
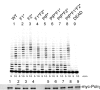
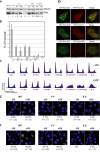
Switching between replicative and translesion synthesis (TLS) DNA polymerases are crucial events for the completion of genomic DNA synthesis when the replication machinery encounters lesions in the DNA template. In eukaryotes, the translesional DNA polymerase η (Polη) plays a central role for accurate bypass of cyclobutane pyrimidine dimers, the predominant DNA lesions induced by ultraviolet irradiation. Polη deficiency is responsible for a variant form of the Xeroderma pigmentosum (XPV) syndrome, characterized by a predisposition to skin cancer. Here, we show that the FF483-484 amino acids in the human Polη (designated F1 motif) are necessary for the interaction of this TLS polymerase with POLD2, the B subunit of the replicative DNA polymerase δ, both in vitro and in vivo. Mutating this motif impairs Polη function in the bypass of both an N-2-acetylaminofluorene adduct and a TT-CPD lesion in cellular extracts. By complementing XPV cells with different forms of Polη, we show that the F1 motif contributes to the progression of DNA synthesis and to the cell survival after UV irradiation. We propose that the integrity of the F1 motif of Polη, necessary for the Polη/POLD2 interaction, is required for the establishment of an efficient TLS complex.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Hoege C., Pfander B., Moldovan G.L., Pyrowolakis G., Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. - PubMed
-
- Stelter P., Ulrich H.D. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–191. - PubMed
-
- Kannouche P.L., Wing J., Lehmann A.R. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell. Biol. 2004;14:491–500. - PubMed
-
- Bienko M., Green C.M., Crosetto N., Rudolf F., Zapart G., Coull B., Kannouche P., Wider G., Peter M., Lehmann A.R., et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310:1821–1824. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

