The PRMT5 arginine methyltransferase: many roles in development, cancer and beyond
- PMID: 25662273
- PMCID: PMC4430368
- DOI: 10.1007/s00018-015-1847-9
The PRMT5 arginine methyltransferase: many roles in development, cancer and beyond
Abstract
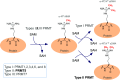
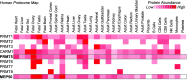
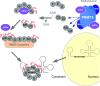
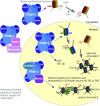
Post-translational arginine methylation is responsible for regulation of many biological processes. The protein arginine methyltransferase 5 (PRMT5, also known as Hsl7, Jbp1, Skb1, Capsuleen, or Dart5) is the major enzyme responsible for mono- and symmetric dimethylation of arginine. An expanding literature demonstrates its critical biological function in a wide range of cellular processes. Histone and other protein methylation by PRMT5 regulate genome organization, transcription, stem cells, primordial germ cells, differentiation, the cell cycle, and spliceosome assembly. Metazoan PRMT5 is found in complex with the WD-repeat protein MEP50 (also known as Wdr77, androgen receptor coactivator p44, or Valois). PRMT5 also directly associates with a range of other protein factors, including pICln, Menin, CoPR5 and RioK1 that may alter its subcellular localization and protein substrate selection. Protein substrate and PRMT5-MEP50 post-translation modifications induce crosstalk to regulate PRMT5 activity. Crystal structures of C. elegans PRMT5 and human and frog PRMT5-MEP50 complexes provide substantial insight into the mechanisms of substrate recognition and procession to dimethylation. Enzymological studies of PRMT5 have uncovered compelling insights essential for future development of specific PRMT5 inhibitors. In addition, newly accumulating evidence implicates PRMT5 and MEP50 expression levels and their methyltransferase activity in cancer tumorigenesis, and, significantly, as markers of poor clinical outcome, marking them as potential oncogenes. Here, we review the substantial new literature on PRMT5 and its partners to highlight the significance of understanding this essential enzyme in health and disease.
Figures


References
-
- Migliori V, Muller J, Phalke S, Low D, Bezzi M, Mok WC, Sahu SK, Gunaratne J, Capasso P, Bassi C, Cecatiello V, De Marco A, Blackstock W, Kuznetsov V, Amati B, Mapelli M, Guccione E. Symmetric dimethylation of H3R2 is a newly identified histone mark that supports euchromatin maintenance. Nat Struct Mol Biol. 2012;19(2):136–144. doi: 10.1038/nsmb.2209. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

