High-throughput and quantitative approaches for measuring circadian rhythms in cyanobacteria using bioluminescence
- PMID: 25662451
- PMCID: PMC4771492
- DOI: 10.1016/bs.mie.2014.10.010
High-throughput and quantitative approaches for measuring circadian rhythms in cyanobacteria using bioluminescence
Abstract
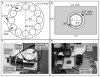
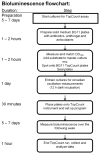
The temporal measurement of a bioluminescent reporter has proven to be one of the most powerful tools for characterizing circadian rhythms in the cyanobacterium Synechococcus elongatus. Primarily, two approaches have been used to automate this process: (1) detection of cell culture bioluminescence in 96-well plates by a photomultiplier tube-based plate-cycling luminometer (TopCount Microplate Scintillation and Luminescence Counter, Perkin Elmer) and (2) detection of individual colony bioluminescence by iteratively rotating a Petri dish under a cooled CCD camera using a computer-controlled turntable. Each approach has distinct advantages. The TopCount provides a more quantitative measurement of bioluminescence, enabling the direct comparison of clock output levels among strains. The computer-controlled turntable approach has a shorter set-up time and greater throughput, making it a more powerful phenotypic screening tool. While the latter approach is extremely useful, only a few labs have been able to build such an apparatus because of technical hurdles involved in coordinating and controlling both the camera and the turntable, and in processing the resulting images. This protocol provides instructions on how to construct, use, and process data from a computer-controlled turntable to measure the temporal changes in bioluminescence of individual cyanobacterial colonies. Furthermore, we describe how to prepare samples for use with the TopCount to minimize experimental noise and generate meaningful quantitative measurements of clock output levels for advanced analysis.
Keywords: Kai proteins; Luciferase; Single colony bioluminescence; Synechococcus elongatus; Temporal automated measurement.
© 2015 Elsevier Inc. All rights reserved.
Figures



References
-
- Andersson C, Tsinoremas N, Shelton J, Lebedeva N, Yarrow J, Min H, et al. Application of bioluminescence to the study of circadian rhythms in cyanobacteria. Methods in Enzymology. 2000;305:527–542. - PubMed
-
- Holtman CK, Chen Y, Sandoval P, Gonzales A, Nalty MS, Thomas TL, et al. High-throughput functional analysis of the Synechococcus elongatus pcc 7942 genome. DNA Research. 2005;12(2):103–115. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

