Sulfur isotope fractionation during the evolutionary adaptation of a sulfate-reducing bacterium
- PMID: 25662968
- PMCID: PMC4375314
- DOI: 10.1128/AEM.03476-14
Sulfur isotope fractionation during the evolutionary adaptation of a sulfate-reducing bacterium
Abstract
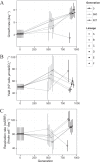
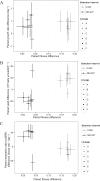
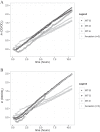
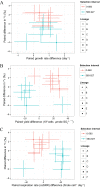
Dissimilatory sulfate reduction is a microbial catabolic pathway that preferentially processes less massive sulfur isotopes relative to their heavier counterparts. This sulfur isotope fractionation is recorded in ancient sedimentary rocks and generally is considered to reflect a phenotypic response to environmental variations rather than to evolutionary adaptation. Modern sulfate-reducing microorganisms isolated from similar environments can exhibit a wide range of sulfur isotope fractionations, suggesting that adaptive processes influence the sulfur isotope phenotype. To date, the relationship between evolutionary adaptation and isotopic phenotypes has not been explored. We addressed this by studying the covariation of fitness, sulfur isotope fractionation, and growth characteristics in Desulfovibrio vulgaris Hildenborough in a microbial evolution experiment. After 560 generations, the mean fitness of the evolved lineages relative to the starting isogenic population had increased by ∼ 17%. After 927 generations, the mean fitness relative to the initial ancestral population had increased by ∼ 20%. Growth rate in exponential phase increased during the course of the experiment, suggesting that this was a primary influence behind the fitness increases. Consistent changes were observed within different selection intervals between fractionation and fitness. Fitness changes were associated with changes in exponential growth rate but changes in fractionation were not. Instead, they appeared to be a response to changes in the parameters that govern growth rate: yield and cell-specific sulfate respiration rate. We hypothesize that cell-specific sulfate respiration rate, in particular, provides a bridge that allows physiological controls on fractionation to cross over to the adaptive realm.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures


References
-
- Minz D, Fishbain S, Green SJ, Muyzer G, Cohen Y, Rittmann BE, Stahl DA. 1999. Unexpected population distribution in a microbial mat community: sulfate-reducing bacteria localized to the highly oxic chemocline in contrast to a eukaryotic preference for anoxia. Appl Environ Microbiol 65:4659–4665. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

