Modeling Viral Capsid Assembly
- PMID: 25663722
- PMCID: PMC4318123
- DOI: 10.1002/9781118755815.ch01
Modeling Viral Capsid Assembly
Abstract
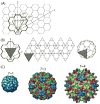
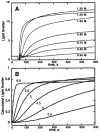
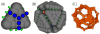
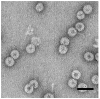
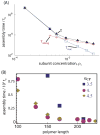
I present a review of the theoretical and computational methodologies that have been used to model the assembly of viral capsids. I discuss the capabilities and limitations of approaches ranging from equilibrium continuum theories to molecular dynamics simulations, and I give an overview of some of the important conclusions about virus assembly that have resulted from these modeling efforts. Topics include the assembly of empty viral shells, assembly around single-stranded nucleic acids to form viral particles, and assembly around synthetic polymers or charged nanoparticles for nanotechnology or biomedical applications. I present some examples in which modeling efforts have promoted experimental breakthroughs, as well as directions in which the connection between modeling and experiment can be strengthened.
Figures









Similar articles
-
Mechanisms of capsid assembly around a polymer.Biophys J. 2010 Jul 21;99(2):619-28. doi: 10.1016/j.bpj.2010.04.035. Biophys J. 2010. PMID: 20643082 Free PMC article.
-
A theory for viral capsid assembly around electrostatic cores.J Chem Phys. 2009 Mar 21;130(11):114902. doi: 10.1063/1.3086041. J Chem Phys. 2009. PMID: 19317561 Free PMC article.
-
Encapsulation of a polymer by an icosahedral virus.Phys Biol. 2010 Dec 9;7(4):045003. doi: 10.1088/1478-3975/7/4/045003. Phys Biol. 2010. PMID: 21149971 Free PMC article.
-
Assembly, Engineering and Applications of Virus-Based Protein Nanoparticles.Adv Exp Med Biol. 2016;940:83-120. doi: 10.1007/978-3-319-39196-0_5. Adv Exp Med Biol. 2016. PMID: 27677510 Review.
-
Mechanisms of virus assembly.Annu Rev Phys Chem. 2015 Apr;66:217-39. doi: 10.1146/annurev-physchem-040214-121637. Epub 2014 Dec 17. Annu Rev Phys Chem. 2015. PMID: 25532951 Free PMC article. Review.
Cited by
-
Single-particle studies of the effects of RNA-protein interactions on the self-assembly of RNA virus particles.Proc Natl Acad Sci U S A. 2022 Sep 27;119(39):e2206292119. doi: 10.1073/pnas.2206292119. Epub 2022 Sep 19. Proc Natl Acad Sci U S A. 2022. PMID: 36122222 Free PMC article.
-
Molecular determinants of Ebola nucleocapsid stability from molecular dynamics simulations.J Chem Phys. 2020 Oct 21;153(15):155102. doi: 10.1063/5.0021491. J Chem Phys. 2020. PMID: 33092380 Free PMC article.
-
Gaussian curvature and the budding kinetics of enveloped viruses.PLoS Comput Biol. 2019 Aug 21;15(8):e1006602. doi: 10.1371/journal.pcbi.1006602. eCollection 2019 Aug. PLoS Comput Biol. 2019. PMID: 31433804 Free PMC article.
-
Thermodynamic Size Control in Curvature-Frustrated Tubules: Self-Limitation with Open Boundaries.ACS Nano. 2022 Jun 28;16(6):9077-9085. doi: 10.1021/acsnano.2c00865. Epub 2022 May 31. ACS Nano. 2022. PMID: 35638478 Free PMC article.
-
Stochastic yield catastrophes and robustness in self-assembly.Elife. 2020 Feb 5;9:e51020. doi: 10.7554/eLife.51020. Elife. 2020. PMID: 32022683 Free PMC article.
References
-
- Adolph KW, Butler PJG. Studies on assembly of a spherical plant virus.1. States of aggregation of isolated protein. J Mol Bio. 1974;88(2):327–341. - PubMed
-
- Alberts Bruce. Essential cell biology. Garland Science; New York; London: 2010.
-
- Alder BJ, Wainwright TE. Studies in molecular dynamics.1. General method. J Chem Phys. 1959;31(2):459–466.
-
- Allen RJ, Frenkel D, ten Wolde PR. Simulating rare events in equilibrium or nonequilibrium stochastic systems. J Chem Phys. 2006;124(2) - PubMed
-
- Allen RJ, Warren PB, ten Wolde PR. Sampling rare switching events in biochemical networks. Phys Rev Lett. 2005;94(1) - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
