Yersinia effector YopO uses actin as bait to phosphorylate proteins that regulate actin polymerization
- PMID: 25664724
- PMCID: PMC4745138
- DOI: 10.1038/nsmb.2964
Yersinia effector YopO uses actin as bait to phosphorylate proteins that regulate actin polymerization
Abstract
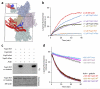
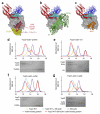
Pathogenic Yersinia species evade host immune systems through the injection of Yersinia outer proteins (Yops) into phagocytic cells. One Yop, YopO, also known as YpkA, induces actin-filament disruption, impairing phagocytosis. Here we describe the X-ray structure of Yersinia enterocolitica YopO in complex with actin, which reveals that YopO binds to an actin monomer in a manner that blocks polymerization yet allows the bound actin to interact with host actin-regulating proteins. SILAC-MS and biochemical analyses confirm that actin-polymerization regulators such as VASP, EVL, WASP, gelsolin and the formin diaphanous 1 are directly sequestered and phosphorylated by YopO through formation of ternary complexes with actin. This leads to a model in which YopO at the membrane sequesters actin from polymerization while using the bound actin as bait to recruit, phosphorylate and misregulate host actin-regulating proteins to disrupt phagocytosis.
Figures




Comment in
-
Subversive bacteria reveal new tricks in their cytoskeleton-hijacking arsenal.Nat Struct Mol Biol. 2015 Mar;22(3):178-9. doi: 10.1038/nsmb.2976. Nat Struct Mol Biol. 2015. PMID: 25736086 Free PMC article.
References
-
- Aderem A, Underhill D. Mechanisms of phagocytosis in macrophages. Annu. Rev. Immunol. 1999;17:593–623. - PubMed
-
- Serrander L, et al. Selective inhibition of IgG-mediated phagocytosis in gelsolin-deficient murine neutrophils. J. Immunol. 2000;165:2451–2457. - PubMed
-
- Colucci-Guyon E, et al. A role for mammalian diaphanous-related formins in complement receptor (CR3)-mediated phagocytosis in macrophages. Curr. Biol. 2005;15:2007–2012. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

