The genomic and phenotypic diversity of Schizosaccharomyces pombe
- PMID: 25665008
- PMCID: PMC4645456
- DOI: 10.1038/ng.3215
The genomic and phenotypic diversity of Schizosaccharomyces pombe
Abstract
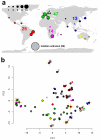
Natural variation within species reveals aspects of genome evolution and function. The fission yeast Schizosaccharomyces pombe is an important model for eukaryotic biology, but researchers typically use one standard laboratory strain. To extend the usefulness of this model, we surveyed the genomic and phenotypic variation in 161 natural isolates. We sequenced the genomes of all strains, finding moderate genetic diversity (π = 3 × 10(-3) substitutions/site) and weak global population structure. We estimate that dispersal of S. pombe began during human antiquity (∼340 BCE), and ancestors of these strains reached the Americas at ∼1623 CE. We quantified 74 traits, finding substantial heritable phenotypic diversity. We conducted 223 genome-wide association studies, with 89 traits showing at least one association. The most significant variant for each trait explained 22% of the phenotypic variance on average, with indels having larger effects than SNPs. This analysis represents a rich resource to examine genotype-phenotype relationships in a tractable model.
Figures

References
-
- Gomes FCO, et al. Physiological diversity and trehalose accumulation in Schizosaccharomyces pombe strains isolated from spontaneous fermentations during the production of the artisanal Brazilian cachaça. Can J Microbiol. 2002;48:399–406. - PubMed
-
- Osterwalder A. Schizosaccharomyces liquefaciens n.sp., eine gegen freie schweflige Säure widerstandsfähige Gärhefe. Mitt Geb Lebensmittelunters Hyg. 1924;15:5–28.
-
- Florenzano G, Balloni W, Materassi R. Contributo alla ecologia dei lieviti Schizosaccharomyces sulle uve. Vitis. 1977;16:38–44.
Methods only references
Publication types
MeSH terms
Associated data
- dbSNP/974514578
- dbSNP/974514579
- dbSNP/974514580
- dbSNP/974514581
- dbSNP/974514582
- dbSNP/974514583
- dbSNP/974514584
- dbSNP/974514585
- dbSNP/974514586
- dbSNP/974514587
- dbSNP/974514588
- dbSNP/974514589
- dbSNP/974514590
- dbSNP/974514591
- dbSNP/974514592
- dbSNP/974514593
- dbSNP/974514594
- dbSNP/974514595
- dbSNP/974514596
- dbSNP/974514597
- dbSNP/974514598
- dbSNP/974514599
- dbSNP/974514600
- dbSNP/974514601
- dbSNP/974514602
- dbSNP/974514603
- dbSNP/974514604
- dbSNP/974514605
- dbSNP/974514606
- dbSNP/974514607
- dbSNP/974514608
- dbSNP/974514609
- dbSNP/974514610
- dbSNP/974514611
- dbSNP/974514612
- dbSNP/974514613
- dbSNP/974514614
- dbSNP/974514615
- dbSNP/974514616
- dbSNP/974514617
- dbSNP/974514618
- dbSNP/974514619
- dbSNP/974514620
- dbSNP/974514621
- dbSNP/974514622
- dbSNP/974514623
- dbSNP/974514624
- dbSNP/974514625
- dbSNP/974514626
- dbSNP/974514627
- dbSNP/974514628
- dbSNP/974514629
- dbSNP/974514630
- dbSNP/974514631
- dbSNP/974514632
- dbSNP/974514633
- dbSNP/974514634
- dbSNP/974514635
- dbSNP/974514636
- dbSNP/974514637
- dbSNP/974514638
- dbSNP/974514639
- dbSNP/974514640
- dbSNP/974514641
- dbSNP/974514642
- dbSNP/974514643
- dbSNP/974514644
- dbSNP/974514645
- dbSNP/974514646
- dbSNP/974514647
- dbSNP/974514648
- dbSNP/974514649
- dbSNP/974514650
- dbSNP/974514651
- dbSNP/974514652
- dbSNP/974514653
- dbSNP/974514654
- dbSNP/974514655
- dbSNP/974514656
- dbSNP/974514657
- dbSNP/974514658
- dbSNP/974514659
- dbSNP/974514660
- dbSNP/974514661
- dbSNP/974514662
- dbSNP/974514663
- dbSNP/974514664
- dbSNP/974514665
- dbSNP/974514666
- dbSNP/974514667
- dbSNP/974514668
- dbSNP/974514669
- dbSNP/974514670
- dbSNP/974514671
- dbSNP/974514672
- dbSNP/974514673
- dbSNP/974514674
- dbSNP/974514675
- dbSNP/974514676
- dbSNP/974514677
- dbSNP/974514678
- dbSNP/974514679
- dbSNP/974514680
- dbSNP/974514681
- dbSNP/974514682
- dbSNP/974514683
- dbSNP/974514684
- dbSNP/974514685
- dbSNP/974514686
- dbSNP/974514687
- dbSNP/974514688
- dbSNP/974514689
- dbSNP/974514690
- dbSNP/974514691
- dbSNP/974514692
- dbSNP/974514693
- dbSNP/974514694
- dbSNP/974514695
- dbSNP/974514696
- dbSNP/974514697
- dbSNP/974514698
- dbSNP/974514699
- dbSNP/974514700
- dbSNP/974514701
- dbSNP/974514702
- dbSNP/974514703
- dbSNP/974514704
- dbSNP/974514705
- dbSNP/974514706
- dbSNP/974514707
- dbSNP/974514708
- dbSNP/974514709
- dbSNP/974514710
- dbSNP/974514711
- dbSNP/974514712
- dbSNP/974514713
- dbSNP/974514714
- dbSNP/974514715
- dbSNP/974514716
- dbSNP/974514717
- dbSNP/974514718
- dbSNP/974514719
- dbSNP/974514720
- dbSNP/974514721
- dbSNP/974514722
- dbSNP/974514723
- dbSNP/974514724
- dbSNP/974514725
- dbSNP/974514726
- dbSNP/974514727
- dbSNP/974514728
- dbSNP/974514729
- dbSNP/974514730
- dbSNP/974514731
- dbSNP/974514732
- dbSNP/974514733
- dbSNP/974514734
- dbSNP/974514735
- dbSNP/974514736
- dbSNP/974514737
- dbSNP/974514738
- dbSNP/974514739
- dbSNP/974514740
- dbSNP/974514741
- dbSNP/974514742
- dbSNP/974514743
- dbSNP/974514744
- dbSNP/974514745
- dbSNP/974514746
- dbSNP/974514747
- dbSNP/974514748
- dbSNP/974514749
- dbSNP/974514750
- dbSNP/974514751
- dbSNP/974514752
- dbSNP/974514753
- dbSNP/974514754
- dbSNP/974514755
- dbSNP/974514756
- dbSNP/974514757
- dbSNP/974514758
- dbSNP/974514759
- dbSNP/974514760
- dbSNP/974514761
- dbSNP/974514762
- dbSNP/974514763
- dbSNP/974514764
- dbSNP/974514765
- dbSNP/974514766
- dbSNP/974514767
- dbSNP/974514768
- dbSNP/974514769
- dbSNP/974514770
- dbSNP/974514771
- dbSNP/974514772
- dbSNP/974514773
- dbSNP/974514774
- dbSNP/974514775
- dbSNP/974514776
- dbSNP/974514777
- dbSNP/974514778
- dbSNP/974514779
- dbSNP/974514780
- dbSNP/974514781
- dbSNP/974514782
- dbSNP/974514783
- dbSNP/974514784
- dbSNP/974514785
- dbSNP/974514786
- dbSNP/974514787
- dbSNP/974514788
- dbSNP/974514789
- dbSNP/974514790
- dbSNP/974514791
- dbSNP/974514792
- dbSNP/974514793
- dbSNP/974514794
- dbSNP/974514795
- dbSNP/974514796
- dbSNP/974514797
- dbSNP/974514798
- dbSNP/974514799
- dbSNP/974514800
- dbSNP/974514801
- dbSNP/974514802
- dbSNP/974514803
- dbSNP/974514804
- dbSNP/974514805
- dbSNP/974514806
- dbSNP/974514807
- dbSNP/974514808
- dbSNP/974514809
- dbSNP/974514810
- dbSNP/974514811
- dbSNP/974514812
- dbSNP/974514813
- dbSNP/974514814
- dbSNP/974514815
- dbSNP/974514816
- dbSNP/974514817
- dbSNP/974514818
- dbSNP/974514819
- dbSNP/974514820
- dbSNP/974514821
- dbSNP/974514822
- dbSNP/974514823
- dbSNP/974514824
- dbSNP/974514825
- dbSNP/974514826
- dbSNP/974514827
- dbSNP/974514828
- dbSNP/974514829
- dbSNP/974514830
- dbSNP/974514831
- dbSNP/974514832
- dbSNP/974514833
- dbSNP/974514834
- dbSNP/974514835
- dbSNP/974514836
- dbSNP/974514837
- dbSNP/974514838
- dbSNP/974514839
- dbSNP/974514840
- dbSNP/974514841
- dbSNP/974514842
- dbSNP/974514843
- dbSNP/974514844
- dbSNP/974514845
- dbSNP/974514846
- dbSNP/974514847
- dbSNP/974514848
- dbSNP/974514849
- dbSNP/974514850
- dbSNP/974514851
- dbSNP/974514852
- dbSNP/974514853
- dbSNP/974514854
- dbSNP/974514855
- dbSNP/974514856
- dbSNP/974514857
- dbSNP/974514858
- dbSNP/974514859
- dbSNP/974514860
- dbSNP/974514861
- dbSNP/974514862
- dbSNP/974514863
- dbSNP/974514864
- dbSNP/974514865
- dbSNP/974514866
- dbSNP/974514867
- dbSNP/974514868
- dbSNP/974514869
- dbSNP/974514870
- dbSNP/974514871
- dbSNP/974514872
- dbSNP/974514873
- dbSNP/974514874
- dbSNP/974514875
- dbSNP/974514876
- dbSNP/974514877
- dbSNP/974514878
- dbSNP/974514879
- dbSNP/974514880
- dbSNP/974514881
- dbSNP/974514882
- dbSNP/974514883
- dbSNP/974514884
- dbSNP/974514885
- dbSNP/974514886
- dbSNP/974514887
- dbSNP/974514888
- dbSNP/974514889
- dbSNP/974514890
- dbSNP/974514891
- dbSNP/974514892
- dbSNP/974514893
- dbSNP/974514894
- dbSNP/974514895
- dbSNP/974514896
- dbSNP/974514897
- dbSNP/974514898
- dbSNP/974514899
- dbSNP/974514900
- dbSNP/974514901
- dbSNP/974514902
- dbSNP/974514903
- dbSNP/974514904
- dbSNP/974514905
- dbSNP/974514906
- dbSNP/974514907
- dbSNP/974514908
- dbSNP/974514909
- dbSNP/974514910
- dbSNP/974514911
- dbSNP/974514912
- dbSNP/974514913
- dbSNP/974514914
- dbSNP/974514915
- dbSNP/974514916
- dbSNP/974514917
- dbSNP/974514918
- dbSNP/974514919
- dbSNP/974514920
- dbSNP/974514921
- dbSNP/974514922
- dbSNP/974514923
- dbSNP/974514924
- dbSNP/974514925
- dbSNP/974514926
- dbSNP/974514927
- dbSNP/974514928
- dbSNP/974514929
- dbSNP/974514930
- dbSNP/974514931
- dbSNP/974514932
- dbSNP/974514933
- dbSNP/974514934
- dbSNP/974514935
- dbSNP/974514936
- dbSNP/974514937
- dbSNP/974514938
- dbSNP/974514939
- dbSNP/974514940
- dbSNP/974514941
- dbSNP/974514942
- dbSNP/974514943
- dbSNP/974514944
- dbSNP/974514945
- dbSNP/974514946
- dbSNP/974514947
- dbSNP/974514948
- dbSNP/974514949
- dbSNP/974514950
- dbSNP/974514951
- dbSNP/974514952
- dbSNP/974514953
- dbSNP/974514954
- dbSNP/974514955
- dbSNP/974514956
- dbSNP/974514957
- dbSNP/974514958
- dbSNP/974514959
- dbSNP/974514960
- dbSNP/974514961
- dbSNP/974514962
- dbSNP/974514963
- dbSNP/974514964
- dbSNP/974514965
- dbSNP/974514966
- dbSNP/974514967
- dbSNP/974514968
- dbSNP/974514969
- dbSNP/974514970
- dbSNP/974514971
- dbSNP/974514972
- dbSNP/974514973
- dbSNP/974514974
- dbSNP/974514975
- dbSNP/974514976
- dbSNP/974514977
- dbSNP/974514978
- dbSNP/974514979
- dbSNP/974514980
- dbSNP/974514981
- dbSNP/974514982
- dbSNP/974514983
- dbSNP/974514984
- dbSNP/974514985
- dbSNP/974514986
- dbSNP/974514987
- dbSNP/974514988
- dbSNP/974514989
- dbSNP/974514990
- dbSNP/974514991
- dbSNP/974514992
- dbSNP/974514993
- dbSNP/974514994
- dbSNP/974514995
- dbSNP/974514996
- dbSNP/974514997
- dbSNP/974514998
- dbSNP/974514999
- dbSNP/974515000
- dbSNP/974515001
- dbSNP/974515002
- dbSNP/974515003
- dbSNP/974515004
- dbSNP/974515005
- dbSNP/974515006
- dbSNP/974515007
- dbSNP/974515008
- dbSNP/974515009
- dbSNP/974515010
- dbSNP/974515011
- dbSNP/974515012
- dbSNP/974515013
- dbSNP/974515014
- dbSNP/974515015
- dbSNP/974515016
- dbSNP/974515017
- dbSNP/974515018
- dbSNP/974515019
- dbSNP/974515020
- dbSNP/974515021
- dbSNP/974515022
- dbSNP/974515023
- dbSNP/974515024
- dbSNP/974515025
- dbSNP/974515026
- dbSNP/974515027
- dbSNP/974515028
- dbSNP/974515029
- dbSNP/974515030
- dbSNP/974515031
- dbSNP/974515032
- dbSNP/974515033
- dbSNP/974515034
- dbSNP/974515035
- dbSNP/974515036
- dbSNP/974515037
- dbSNP/974515038
- dbSNP/974515039
- dbSNP/974515040
- dbSNP/974515041
- dbSNP/974515042
- dbSNP/974515043
- dbSNP/974515044
- dbSNP/974515045
- dbSNP/974515046
- dbSNP/974515047
- dbSNP/974515048
- dbSNP/974515049
- dbSNP/974515050
- dbSNP/974515051
- dbSNP/974515052
- dbSNP/974515053
- dbSNP/974515054
- dbSNP/974515055
- dbSNP/974515056
- dbSNP/974515057
- dbSNP/974515058
- dbSNP/974515059
- dbSNP/974515060
- dbSNP/974515061
- dbSNP/974515062
- dbSNP/974515063
- dbSNP/974515064
- dbSNP/974515065
- dbSNP/974515066
- dbSNP/974515067
- dbSNP/974515068
- dbSNP/974515069
- dbSNP/974515070
- dbSNP/974515071
- dbSNP/974515072
- dbSNP/974515073
- dbSNP/974515074
- dbSNP/974515075
- dbSNP/974515076
- dbSNP/974515077
- dbSNP/974515078
- dbSNP/974515079
- dbSNP/974515080
- dbSNP/974515081
- dbSNP/974515082
- dbSNP/974515083
- dbSNP/974515084
- dbSNP/974515085
- dbSNP/974515086
- dbSNP/974515087
- dbSNP/974515088
- dbSNP/974515089
- dbSNP/974515090
- dbSNP/974515091
- dbSNP/974515092
- dbSNP/974515093
- dbSNP/974515094
- dbSNP/974515095
- dbSNP/974515096
- dbSNP/974515097
- dbSNP/974515098
- dbSNP/974515099
- dbSNP/974515100
- dbSNP/974515101
- dbSNP/974515102
- dbSNP/974515103
- dbSNP/974515104
- dbSNP/974515105
- dbSNP/974515106
- dbSNP/974515107
- dbSNP/974515108
- dbSNP/974515109
- dbSNP/974515110
- dbSNP/974515111
- dbSNP/974515112
- dbSNP/974515113
- dbSNP/974515114
- dbSNP/974515115
- dbSNP/974515116
- dbSNP/974515117
- dbSNP/974515118
- dbSNP/974515119
- dbSNP/974515120
- dbSNP/974515121
- dbSNP/974515122
- dbSNP/974515123
- dbSNP/974515124
- dbSNP/974515125
- dbSNP/974515126
- dbSNP/974515127
- dbSNP/974515128
- dbSNP/974515129
- dbSNP/974515130
- dbSNP/974515131
- dbSNP/974515132
- dbSNP/974515133
- dbSNP/974515134
- dbSNP/974515135
- dbSNP/974515136
- dbSNP/974515137
- dbSNP/974515138
- dbSNP/974515139
- dbSNP/974515140
- dbSNP/974515141
- dbSNP/974515142
- dbSNP/974515143
- dbSNP/974515144
- dbSNP/974515145
- dbSNP/974515146
- dbSNP/974515147
- dbSNP/974515148
- dbSNP/974515149
- dbSNP/974515150
- dbSNP/974515151
- dbSNP/974515152
- dbSNP/974515153
- dbSNP/974515154
- dbSNP/974515155
- dbSNP/974515156
- dbSNP/974515157
- dbSNP/974515158
- dbSNP/974515159
- dbSNP/974515160
- dbSNP/974515161
- dbSNP/974515162
- dbSNP/974515163
- dbSNP/974515164
- dbSNP/974515165
- dbSNP/974515166
- dbSNP/974515167
- dbSNP/974515168
- dbSNP/974515169
- dbSNP/974515170
- dbSNP/974515171
- dbSNP/974515172
- dbSNP/974515173
- dbSNP/974515174
- dbSNP/974515175
- dbSNP/974515176
- dbSNP/974515177
- dbSNP/974515178
- dbSNP/974515179
- dbSNP/974515180
- dbSNP/974515181
- dbSNP/974515182
- dbSNP/974515183
- dbSNP/974515184
- dbSNP/974515185
- dbSNP/974515186
- dbSNP/974515187
- dbSNP/974515188
- dbSNP/974515189
- dbSNP/974515190
- dbSNP/974515191
- dbSNP/974515192
- dbSNP/974515193
- dbSNP/974515194
- dbSNP/974515195
- dbSNP/974515196
- dbSNP/974515197
- dbSNP/974515198
- dbSNP/974515199
- dbSNP/974515200
- dbSNP/974515201
- dbSNP/974515202
- dbSNP/974515203
- dbSNP/974515204
- dbSNP/974515205
- dbSNP/974515206
- dbSNP/974515207
- dbSNP/974515208
- dbSNP/974515209
- dbSNP/974515210
- dbSNP/974515211
- dbSNP/974515212
- dbSNP/974515213
- dbSNP/974515214
- dbSNP/974515215
- dbSNP/974515216
- dbSNP/974515217
- dbSNP/974515218
- dbSNP/974515219
- dbSNP/974515220
- dbSNP/974515221
- dbSNP/974515222
- dbSNP/974515223
- dbSNP/974515224
- dbSNP/974515225
- dbSNP/974515226
- dbSNP/974515227
- dbSNP/974515228
- dbSNP/974515229
- dbSNP/974515230
- dbSNP/974515231
- dbSNP/974515232
- dbSNP/974515233
- dbSNP/974515234
- dbSNP/974515235
- dbSNP/974515236
- dbSNP/974515237
- dbSNP/974515238
- dbSNP/974515239
- dbSNP/974515240
- dbSNP/974515241
- dbSNP/974515242
- dbSNP/974515243
- dbSNP/974515244
- dbSNP/974515245
- dbSNP/974515246
- dbSNP/974515247
- dbSNP/974515248
- dbSNP/974515249
- dbSNP/974515250
- dbSNP/974515251
- dbSNP/974515252
- dbSNP/974515253
- dbSNP/974515254
- dbSNP/974515255
- dbSNP/974515256
- dbSNP/974515257
- dbSNP/974515258
- dbSNP/974515259
- dbSNP/974515260
- dbSNP/974515261
- dbSNP/974515262
- dbSNP/974515263
- dbSNP/974515264
- dbSNP/974515265
- dbSNP/974515266
- dbSNP/974515267
- dbSNP/974515268
- dbSNP/974515269
- dbSNP/974515270
- dbSNP/974515271
- dbSNP/974515272
- dbSNP/974515273
- dbSNP/974515274
- dbSNP/974515275
- dbSNP/974515276
- dbSNP/974515277
- dbSNP/974515278
- dbSNP/974515279
- dbSNP/974515280
- dbSNP/974515281
- dbSNP/974515282
- dbSNP/974515283
- dbSNP/974515284
- dbSNP/974515285
- dbSNP/974515286
- dbSNP/974515287
- dbSNP/974515288
- dbSNP/974515289
- dbSNP/974515290
- dbSNP/974515291
- dbSNP/974515292
- dbSNP/974515293
- dbSNP/974515294
- dbSNP/974515295
- dbSNP/974515296
- dbSNP/974515297
- dbSNP/974515298
- dbSNP/974515299
- dbSNP/974515300
- dbSNP/974515301
- dbSNP/974515302
- dbSNP/974515303
- dbSNP/974515304
- dbSNP/974515305
- dbSNP/974515306
- dbSNP/974515307
- dbSNP/974515308
- dbSNP/974515309
- dbSNP/974515310
- dbSNP/974515311
- dbSNP/974515312
- dbSNP/974515313
- dbSNP/974515314
- dbSNP/974515315
- dbSNP/974515316
- dbSNP/974515317
- dbSNP/974515318
- dbSNP/974515319
- dbSNP/974515320
- dbSNP/974515321
- dbSNP/974515322
- dbSNP/974515323
- dbSNP/974515324
- dbSNP/974515325
- dbSNP/974515326
- dbSNP/974515327
- dbSNP/974515328
- dbSNP/974515329
- dbSNP/974515330
- dbSNP/974515331
- dbSNP/974515332
- dbSNP/974515333
- dbSNP/974515334
- dbSNP/974515335
- dbSNP/974515336
- dbSNP/974515337
- dbSNP/974515338
- dbSNP/974515339
- dbSNP/974515340
- dbSNP/974515341
- dbSNP/974515342
- dbSNP/974515343
- dbSNP/974515344
- dbSNP/974515345
- dbSNP/974515346
- dbSNP/974515347
- dbSNP/974515348
- dbSNP/974515349
- dbSNP/974515350
- dbSNP/974515351
- dbSNP/974515352
- dbSNP/974515353
- dbSNP/974515354
- dbSNP/974515355
- dbSNP/974515356
- dbSNP/974515357
- dbSNP/974515358
- dbSNP/974515359
- dbSNP/974515360
- dbSNP/974515361
- dbSNP/974515362
- dbSNP/974515363
- dbSNP/974515364
- dbSNP/974515365
- dbSNP/974515366
- dbSNP/974515367
- dbSNP/974515368
- dbSNP/974515369
- dbSNP/974515370
- dbSNP/974515371
- dbSNP/974515372
- dbSNP/974515373
- dbSNP/974515374
- dbSNP/974515375
- dbSNP/974515376
- dbSNP/974515377
- dbSNP/974515378
- dbSNP/974515379
- dbSNP/974515380
- dbSNP/974515381
- dbSNP/974515382
- dbSNP/974515383
- dbSNP/974515384
- dbSNP/974515385
- dbSNP/974515386
- dbSNP/974515387
- dbSNP/974515388
- dbSNP/974515389
- dbSNP/974515390
- dbSNP/974515391
- dbSNP/974515392
- dbSNP/974515393
- dbSNP/974515394
- dbSNP/974515395
- dbSNP/974515396
- dbSNP/974515397
- dbSNP/974515398
- dbSNP/974515399
- dbSNP/974515400
- dbSNP/974515401
- dbSNP/974515402
- dbSNP/974515403
- dbSNP/974515404
- dbSNP/974515405
- dbSNP/974515406
- dbSNP/974515407
- dbSNP/974515408
- dbSNP/974515409
- dbSNP/974515410
- dbSNP/974515411
- dbSNP/974515412
- dbSNP/974515413
- dbSNP/974515414
- dbSNP/974515415
- dbSNP/974515416
- dbSNP/974515417
- dbSNP/974515418
- dbSNP/974515419
- dbSNP/974515420
- dbSNP/974515421
- dbSNP/974515422
- dbSNP/974515423
- dbSNP/974515424
- dbSNP/974515425
- dbSNP/974515426
- dbSNP/974515427
- dbSNP/974515428
- dbSNP/974515429
- dbSNP/974515430
- dbSNP/974515431
- dbSNP/974515432
- dbSNP/974515433
- dbSNP/974515434
- dbSNP/974515435
- dbSNP/974515436
- dbSNP/974515437
- dbSNP/974515438
- dbSNP/974515439
- dbSNP/974515440
- dbSNP/974515441
- dbSNP/974515442
- dbSNP/974515443
- dbSNP/974515444
- dbSNP/974515445
- dbSNP/974515446
- dbSNP/974515447
- dbSNP/974515448
- dbSNP/974515449
- dbSNP/974515450
- dbSNP/974515451
- dbSNP/974515452
- dbSNP/974515453
- dbSNP/974515454
- dbSNP/974515455
- dbSNP/974515456
- dbSNP/974515457
- dbSNP/974515458
- dbSNP/974515459
- dbSNP/974515460
- dbSNP/974515461
- dbSNP/974515462
- dbSNP/974515463
- dbSNP/974515464
- dbSNP/974515465
- dbSNP/974515466
- dbSNP/974515467
- dbSNP/974515468
- dbSNP/974515469
- dbSNP/974515470
- dbSNP/974515471
- dbSNP/974515472
- dbSNP/974515473
- dbSNP/974515474
- dbSNP/974515475
- dbSNP/974515476
- dbSNP/974515477
- dbSNP/974515478
- dbSNP/974515479
- dbSNP/974515480
- dbSNP/974515481
- dbSNP/974515482
- dbSNP/974515483
- dbSNP/974515484
- dbSNP/974515485
- dbSNP/974515486
- dbSNP/974515487
- dbSNP/974515488
- dbSNP/974515489
- dbSNP/974515490
- dbSNP/974515491
- dbSNP/974515492
- dbSNP/974515493
- dbSNP/974515494
- dbSNP/974515495
- dbSNP/974515496
- dbSNP/974515497
- dbSNP/974515498
- dbSNP/974515499
- dbSNP/974515500
- dbSNP/974515501
- dbSNP/974515502
- dbSNP/974515503
- dbSNP/974515504
- dbSNP/974515505
- dbSNP/974515506
- dbSNP/974515507
- dbSNP/974515508
- dbSNP/974515509
- dbSNP/974515510
- dbSNP/974515511
- dbSNP/974515512
- dbSNP/974515513
- dbSNP/974515514
- dbSNP/974515515
- dbSNP/974515516
- dbSNP/974515517
- dbSNP/974515518
- dbSNP/974515519
- dbSNP/974515520
- dbSNP/974515521
- dbSNP/974515522
- dbSNP/974515523
- dbSNP/974515524
- dbSNP/974515525
- dbSNP/974515526
- dbSNP/974515527
- dbSNP/974515528
- dbSNP/974515529
- dbSNP/974515530
- dbSNP/974515531
- dbSNP/974515532
- dbSNP/974515533
- dbSNP/974515534
- dbSNP/974515535
- dbSNP/974515536
- dbSNP/974515537
- dbSNP/974515538
- dbSNP/974515539
- dbSNP/974515540
- dbSNP/974515541
- dbSNP/974515542
- dbSNP/974515543
- dbSNP/974515544
- dbSNP/974515545
- dbSNP/974515546
- dbSNP/974515547
- dbSNP/974515548
- dbSNP/974515549
- dbSNP/974515550
- dbSNP/974515551
- dbSNP/974515552
- dbSNP/974515553
- dbSNP/974515554
- dbSNP/974515555
- dbSNP/974515556
- dbSNP/974515557
- dbSNP/974515558
- dbSNP/974515559
- dbSNP/974515560
- dbSNP/974515561
- dbSNP/974515562
- dbSNP/974515563
- dbSNP/974515564
- dbSNP/974515565
- dbSNP/974515566
- dbSNP/974515567
- dbSNP/974515568
- dbSNP/974515569
- dbSNP/974515570
- dbSNP/974515571
- dbSNP/974515572
- dbSNP/974515573
- dbSNP/974515574
- dbSNP/974515575
- dbSNP/974515576
- dbSNP/974515577
Grants and funding
- 093917/WT_/Wellcome Trust/United Kingdom
- BB/K006320/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 095598/WT_/Wellcome Trust/United Kingdom
- BB/H005854/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 098051/WT_/Wellcome Trust/United Kingdom
- 093735/WT_/Wellcome Trust/United Kingdom
- 260809/ERC_/European Research Council/International
- RG 093735/Z/10/Z/WT_/Wellcome Trust/United Kingdom
- BB/H008802/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 15699/CRUK_/Cancer Research UK/United Kingdom
- G0901388/MRC_/Medical Research Council/United Kingdom
- 095598/Z/11/Z/WT_/Wellcome Trust/United Kingdom
- 260801/ERC_/European Research Council/International
- 098386/WT_/Wellcome Trust/United Kingdom
- MR/L012561/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

