Molecular, genetic, and cellular bases for treating eosinophilic esophagitis
- PMID: 25666870
- PMCID: PMC4409569
- DOI: 10.1053/j.gastro.2015.02.002
Molecular, genetic, and cellular bases for treating eosinophilic esophagitis
Abstract
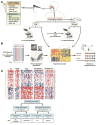
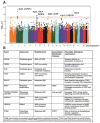
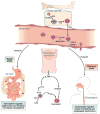
Eosinophilic esophagitis (EoE) was historically distinguished from gastroesophageal reflux disease on the basis of histology and lack of responsiveness to acid suppressive therapy, but it is now appreciated that esophageal eosinophilia can respond to proton pump inhibitors. Genetic and environmental factors contribute to risk for EoE, particularly early-life events. Disease pathogenesis involves activation of epithelial inflammatory pathways (production of eotaxin-3 [encoded by CCL26]), impaired barrier function (mediated by loss of desmoglein-1), increased production and/or activity of transforming growth factor-β, and induction of allergic inflammation by eosinophils and mast cells. Susceptibility has been associated with variants at 5q22 (TSLP) and 2p23 (CAPN14), indicating roles for allergic sensitization and esophageal specific protease pathways. We propose that EoE is a unique disease characterized by food hypersensitivity; strong hereditability influenced by early-life exposures and esophageal-specific genetic risk variants; and allergic inflammation and that the disease is remitted by disrupting inflammatory and T-helper type 2 cytokine-mediated responses and through dietary elimination therapy.
Keywords: Allergy; Barrier; Cytokines; Diagnostics; Eosinophils; Epithelium; Esophagitis; Genetics; Therapy.
Copyright © 2015 AGA Institute. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Liacouras CA, Furuta GT, Hirano I, et al. Eosinophilic esophagitis: updated consensus recommendations for children and adults. J Allergy Clin Immunol. 2011;128:3–20 e6. quiz 21-2. - PubMed
-
- Dellon ES, Gonsalves N, Hirano I, et al. ACG Clinical Guideline: Evidenced Based Approach to the Diagnosis and Management of Esophageal Eosinophilia and Eosinophilic Esophagitis (EoE) Am J Gastroenterol. 2013;108:679–92. - PubMed
-
- Dellon EP, Liacouras C. Eosinophilic Esophagitis. Gastroenterol. 2015 In Press.
-
- Attwood SE, Smyrk TC, Demeester TR, et al. Esophageal eosinophilia with dysphagia. A distinct clinicopathologic syndrome. Dig Dis Sci. 1993;38:109–16. - PubMed
-
- Kelly KJ, Lazenby AJ, Rowe PC, et al. Eosinophilic esophagitis attributed to gastroesophageal reflux: improvement with an amino acid-based formula. Gastroenterology. 1995;109:1503–12. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

