Genotyping coronaviruses associated with feline infectious peritonitis
- PMID: 25667330
- PMCID: PMC4635486
- DOI: 10.1099/vir.0.000084
Genotyping coronaviruses associated with feline infectious peritonitis
Abstract
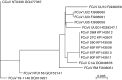
Feline coronavirus (FCoV) infections are endemic among cats worldwide. The majority of infections are asymptomatic or result in only mild enteric disease. However, approximately 5 % of cases develop feline infectious peritonitis (FIP), a systemic disease that is a frequent cause of death in young cats. In this study, we report the complete coding genome sequences of six FCoVs: three from faecal samples from healthy cats and three from tissue lesion samples from cats with confirmed FIP. The six samples were obtained over a period of 8 weeks at a single-site cat rescue and rehoming centre in the UK. We found amino acid differences located at 44 positions across an alignment of the six virus translatomes and, at 21 of these positions, the differences fully or partially discriminated between the genomes derived from the faecal samples and the genomes derived from the tissue lesion samples. In this study, two amino acid differences fully discriminated the two classes of genomes: these were both located in the S2 domain of the virus surface glycoprotein gene. We also identified deletions in the 3c protein ORF of genomes from two of the FIP samples. Our results support previous studies that implicate S protein mutations in the pathogenesis of FIP.
© 2015 The Authors.
Figures




References
-
- Barker E. N., Tasker S., Gruffydd-Jones T. J., Tuplin C. K., Burton K., Porter E., Day M. J., Harley R., Fews D., et al. ( 2013. ). Phylogenetic analysis of feline coronavirus strains in an epizootic outbreak of feline infectious peritonitis. J Vet Intern Med 27, 445–450. 10.1111/jvim.12058 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

