The human urine virome in association with urinary tract infections
- PMID: 25667584
- PMCID: PMC4304238
- DOI: 10.3389/fmicb.2015.00014
The human urine virome in association with urinary tract infections
Abstract
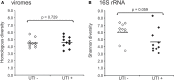
While once believed to represent a sterile environment, the human urinary tract harbors a unique cellular microbiota. We sought to determine whether the human urinary tract also is home to viral communities whose membership might reflect urinary tract health status. We recruited and sampled urine from 20 subjects, 10 subjects with urinary tract infections (UTIs) and 10 without UTIs, and found viral communities in the urine of each subject group. Most of the identifiable viruses were bacteriophage, but eukaryotic viruses also were identified in all subjects. We found reads from human papillomaviruses (HPVs) in 95% of the subjects studied, but none were found to be high-risk genotypes that are associated with cervical and rectal cancers. We verified the presence of some HPV genotypes by quantitative PCR. Some of the HPV genotypes identified were homologous to relatively novel and uncharacterized viruses that previously have been detected on skin in association with cancerous lesions, while others may be associated with anal and genital warts. On a community level, there was no association between the membership or diversity of viral communities based on urinary tract health status. While more data are still needed, detection of HPVs as members of the human urinary virome using viral metagenomics represents a non-invasive technique that could augment current screening techniques to detect low-risk HPVs in the genitourinary tracts of humans.
Keywords: HPV; human microbiome; papillomavirus; urinary tract infections; virobiota; virome.
Figures