Vision from next generation sequencing: multi-dimensional genome-wide analysis for producing gene regulatory networks underlying retinal development, aging and disease
- PMID: 25668385
- PMCID: PMC4402139
- DOI: 10.1016/j.preteyeres.2015.01.005
Vision from next generation sequencing: multi-dimensional genome-wide analysis for producing gene regulatory networks underlying retinal development, aging and disease
Abstract
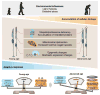
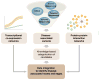
Genomics and genetics have invaded all aspects of biology and medicine, opening uncharted territory for scientific exploration. The definition of "gene" itself has become ambiguous, and the central dogma is continuously being revised and expanded. Computational biology and computational medicine are no longer intellectual domains of the chosen few. Next generation sequencing (NGS) technology, together with novel methods of pattern recognition and network analyses, has revolutionized the way we think about fundamental biological mechanisms and cellular pathways. In this review, we discuss NGS-based genome-wide approaches that can provide deeper insights into retinal development, aging and disease pathogenesis. We first focus on gene regulatory networks (GRNs) that govern the differentiation of retinal photoreceptors and modulate adaptive response during aging. Then, we discuss NGS technology in the context of retinal disease and develop a vision for therapies based on network biology. We should emphasize that basic strategies for network construction and analyses can be transported to any tissue or cell type. We believe that specific and uniform guidelines are required for generation of genome, transcriptome and epigenome data to facilitate comparative analysis and integration of multi-dimensional data sets, and for constructing networks underlying complex biological processes. As cellular homeostasis and organismal survival are dependent on gene-gene and gene-environment interactions, we believe that network-based biology will provide the foundation for deciphering disease mechanisms and discovering novel drug targets for retinal neurodegenerative diseases.
Keywords: ChIP-seq; Gene regulatory network; High throughput genomics; Inherited blindness; Macular degeneration; Network medicine; Pathway-based drug discovery; Personalized medicine; Photoreceptor; RNA-seq; Retinal degeneration; Systems biology; Whole exome sequencing; eQTL.
Published by Elsevier Ltd.
Figures






References
-
- Agathocleous M, Harris WA. From progenitors to differentiated cells in the vertebrate retina. Annu Rev Cell Dev Biol. 2009;25:45–69. - PubMed
-
- Akimoto M, Cheng H, Zhu D, Brzezinski JA, Khanna R, Filippova E, Oh EC, Jing Y, Linares JL, Brooks M, Zareparsi S, Mears AJ, Hero A, Glaser T, Swaroop A. Targeting of GFP to newborn rods by Nrl promoter and temporal expression profiling of flow-sorted photoreceptors. Proc Natl Acad Sci U S A. 2006;103:3890–3895. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

