Systems-based analysis of the Sarcocystis neurona genome identifies pathways that contribute to a heteroxenous life cycle
- PMID: 25670772
- PMCID: PMC4337577
- DOI: 10.1128/mBio.02445-14
Systems-based analysis of the Sarcocystis neurona genome identifies pathways that contribute to a heteroxenous life cycle
Abstract
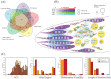
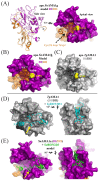
Sarcocystis neurona is a member of the coccidia, a clade of single-celled parasites of medical and veterinary importance including Eimeria, Sarcocystis, Neospora, and Toxoplasma. Unlike Eimeria, a single-host enteric pathogen, Sarcocystis, Neospora, and Toxoplasma are two-host parasites that infect and produce infectious tissue cysts in a wide range of intermediate hosts. As a genus, Sarcocystis is one of the most successful protozoan parasites; all vertebrates, including birds, reptiles, fish, and mammals are hosts to at least one Sarcocystis species. Here we sequenced Sarcocystis neurona, the causal agent of fatal equine protozoal myeloencephalitis. The S. neurona genome is 127 Mbp, more than twice the size of other sequenced coccidian genomes. Comparative analyses identified conservation of the invasion machinery among the coccidia. However, many dense-granule and rhoptry kinase genes, responsible for altering host effector pathways in Toxoplasma and Neospora, are absent from S. neurona. Further, S. neurona has a divergent repertoire of SRS proteins, previously implicated in tissue cyst formation in Toxoplasma. Systems-based analyses identified a series of metabolic innovations, including the ability to exploit alternative sources of energy. Finally, we present an S. neurona model detailing conserved molecular innovations that promote the transition from a purely enteric lifestyle (Eimeria) to a heteroxenous parasite capable of infecting a wide range of intermediate hosts.
Importance: Sarcocystis neurona is a member of the coccidia, a clade of single-celled apicomplexan parasites responsible for major economic and health care burdens worldwide. A cousin of Plasmodium, Cryptosporidium, Theileria, and Eimeria, Sarcocystis is one of the most successful parasite genera; it is capable of infecting all vertebrates (fish, reptiles, birds, and mammals-including humans). The past decade has witnessed an increasing number of human outbreaks of clinical significance associated with acute sarcocystosis. Among Sarcocystis species, S. neurona has a wide host range and causes fatal encephalitis in horses, marine mammals, and several other mammals. To provide insights into the transition from a purely enteric parasite (e.g., Eimeria) to one that forms tissue cysts (Toxoplasma), we present the first genome sequence of S. neurona. Comparisons with other coccidian genomes highlight the molecular innovations that drive its distinct life cycle strategies.
Copyright © 2015 Blazejewski et al.
Figures




References
-
- Dubey JP, Speer CA, Fayer R. 1988. Sarcocystosis of animals and man. CRC Press, Boca Raton, FL.
-
- Wasmuth JD, Pszenny V, Haile S, Jansen EM, Gast AT, Sher A, Boyle JP, Boulanger MJ, Parkinson J, Grigg ME. 2012. Integrated bioinformatic and targeted deletion analyses of the SRS gene superfamily identify SRS29C as a negative regulator of Toxoplasma virulence. mBio 3(6):e00321-12. doi:10.1128/mBio.00321-12. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

