Genomes of planktonic Acidimicrobiales: widening horizons for marine Actinobacteria by metagenomics
- PMID: 25670777
- PMCID: PMC4337565
- DOI: 10.1128/mBio.02083-14
Genomes of planktonic Acidimicrobiales: widening horizons for marine Actinobacteria by metagenomics
Abstract
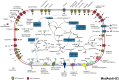
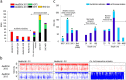
The genomes of four novel marine Actinobacteria have been assembled from large metagenomic data sets derived from the Mediterranean deep chlorophyll maximum (DCM). These are the first marine representatives belonging to the order Acidimicrobiales and only the second group of planktonic marine Actinobacteria to be described. Their streamlined genomes and photoheterotrophic lifestyle suggest that they are planktonic, free-living microbes. A novel rhodopsin clade, acidirhodopsins, related to freshwater actinorhodopsins, was found in these organisms. Their genomes suggest a capacity to assimilate C2 compounds, some using the glyoxylate bypass and others with the ethylmalonyl-coenzyme A (CoA) pathway. They are also able to derive energy from dimethylsulfopropionate (DMSP), sulfonate, and carbon monoxide oxidation, all commonly available in the marine habitat. These organisms appear to be prevalent in the deep photic zone at or around the DCM. The presence of sister clades to the marine Acidimicrobiales in freshwater aquatic habitats provides a new example of marine-freshwater transitions with potential evolutionary insights.
Importance: Despite several studies showing the importance and abundance of planktonic Actinobacteria in the marine habitat, a representative genome was only recently described. In order to expand the genomic repertoire of marine Actinobacteria, we describe here the first Acidimicrobidae genomes of marine origin and provide insights about their ecology. They display metabolic versatility in the acquisition of carbon and appear capable of utilizing diverse sources of energy. One of the genomes harbors a new kind of rhodopsin related to the actinorhodopsin clade of freshwater origin that is widespread in the oceans. Our data also support their preference to inhabit the deep chlorophyll maximum and the deep photic zone. This work contributes to the perception of marine actinobacterial groups as important players in the marine environment with distinct and important contributions to nutrient cycling in the oceans.
Copyright © 2015 Mizuno et al.
Figures



References
-
- Rappé MS, Kemp PF, Giovannoni SJ. 1997. Phylogenetic diversity of marine coastal picoplankton 16S rRNA genes cloned from the continental shelf off Cape Hatteras, North Carolina. Limnol Oceangr 42:811–826. doi:10.4319/lo.1997.42.5.0811. - DOI
-
- Rappé MS, Gordon DA, Vergin KL, Giovannoni SJ. 1999. Phylogeny of Actinobacteria small subunit (SSU) rRNA gene clones recovered from marine bacterioplankton. Syst Appl Microbiol 22:106–112. doi:10.1016/S0723-2020(99)80033-2. - DOI
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous
