Enteroendocrine cells are generated from stem cells through a distinct progenitor in the adult Drosophila posterior midgut
- PMID: 25670791
- PMCID: PMC4325374
- DOI: 10.1242/dev.113357
Enteroendocrine cells are generated from stem cells through a distinct progenitor in the adult Drosophila posterior midgut
Abstract
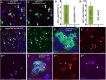
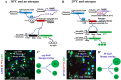
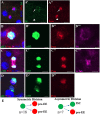
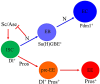
Functional mature cells are continually replenished by stem cells to maintain tissue homoeostasis. In the adult Drosophila posterior midgut, both terminally differentiated enterocyte (EC) and enteroendocrine (EE) cells are generated from an intestinal stem cell (ISC). However, it is not clear how the two differentiated cells are generated from the ISC. In this study, we found that only ECs are generated through the Su(H)GBE(+) immature progenitor enteroblasts (EBs), whereas EEs are generated from ISCs through a distinct progenitor pre-EE by a novel lineage-tracing system. EEs can be generated from ISCs in three ways: an ISC becoming an EE, an ISC becoming a new ISC and an EE through asymmetric division, or an ISC becoming two EEs through symmetric division. We further identified that the transcriptional factor Prospero (Pros) regulates ISC commitment to EEs. Our data provide direct evidence that different differentiated cells are generated by different modes of stem cell lineage specification within the same tissues.
Keywords: Drosophila; Intestinal stem cell; Pre-enteroendocrine cell; Prospero; Secretory enteroendocrine cell.
© 2015. Published by The Company of Biologists Ltd.
Figures



Similar articles
-
Ttk69 acts as a master repressor of enteroendocrine cell specification in Drosophila intestinal stem cell lineages.Development. 2015 Oct 1;142(19):3321-31. doi: 10.1242/dev.123208. Epub 2015 Aug 20. Development. 2015. PMID: 26293304
-
Keeping intestinal stem cell differentiation on the Tramtrack.Fly (Austin). 2015;9(3):110-4. doi: 10.1080/19336934.2015.1119947. Fly (Austin). 2015. PMID: 26655207 Free PMC article.
-
A Phyllopod-Mediated Feedback Loop Promotes Intestinal Stem Cell Enteroendocrine Commitment in Drosophila.Stem Cell Reports. 2018 Jan 9;10(1):43-57. doi: 10.1016/j.stemcr.2017.11.014. Epub 2017 Dec 21. Stem Cell Reports. 2018. PMID: 29276156 Free PMC article.
-
Intestinal stem cell response to injury: lessons from Drosophila.Cell Mol Life Sci. 2016 Sep;73(17):3337-49. doi: 10.1007/s00018-016-2235-9. Epub 2016 May 2. Cell Mol Life Sci. 2016. PMID: 27137186 Free PMC article. Review.
-
Basic helix-loop-helix transcription factors and enteroendocrine cell differentiation.Diabetes Obes Metab. 2011 Oct;13 Suppl 1(0 1):5-12. doi: 10.1111/j.1463-1326.2011.01438.x. Diabetes Obes Metab. 2011. PMID: 21824251 Free PMC article. Review.
Cited by
-
Tissue Adaptation to Environmental Cues by Symmetric and Asymmetric Division Modes of Intestinal Stem Cells.Int J Mol Sci. 2020 Sep 2;21(17):6362. doi: 10.3390/ijms21176362. Int J Mol Sci. 2020. PMID: 32887329 Free PMC article. Review.
-
The Intestinal Stem Cell Niche: Homeostasis and Adaptations.Trends Cell Biol. 2018 Dec;28(12):1062-1078. doi: 10.1016/j.tcb.2018.08.001. Epub 2018 Sep 5. Trends Cell Biol. 2018. PMID: 30195922 Free PMC article. Review.
-
The WT1-like transcription factor Klumpfuss maintains lineage commitment of enterocyte progenitors in the Drosophila intestine.Nat Commun. 2019 Sep 11;10(1):4123. doi: 10.1038/s41467-019-12003-0. Nat Commun. 2019. PMID: 31511511 Free PMC article.
-
Autophagy maintains stem cells and intestinal homeostasis in Drosophila.Sci Rep. 2018 Mar 15;8(1):4644. doi: 10.1038/s41598-018-23065-3. Sci Rep. 2018. PMID: 29545557 Free PMC article.
-
Insect Gut Regeneration.Cold Spring Harb Perspect Biol. 2022 Feb 1;14(2):a040915. doi: 10.1101/cshperspect.a040915. Cold Spring Harb Perspect Biol. 2022. PMID: 34312250 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

