Haploinsufficiency of the c-myc transcriptional repressor FIR, as a dominant negative-alternative splicing model, promoted p53-dependent T-cell acute lymphoblastic leukemia progression by activating Notch1
- PMID: 25671302
- PMCID: PMC4467136
- DOI: 10.18632/oncotarget.3244
Haploinsufficiency of the c-myc transcriptional repressor FIR, as a dominant negative-alternative splicing model, promoted p53-dependent T-cell acute lymphoblastic leukemia progression by activating Notch1
Abstract
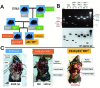
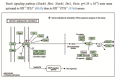
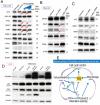
FUSE-binding protein (FBP)-interacting repressor (FIR) is a c-myc transcriptional suppressor. A splice variant of FIR that lacks exon 2 in the transcriptional repressor domain (FIRΔexon2) upregulates c-myc transcription by inactivating wild-type FIR. The ratio of FIRΔexon2/FIR mRNA was increased in human colorectal cancer and hepatocellular carcinoma tissues. Because FIRΔexon2 is considered to be a dominant negative regulator of FIR, FIR heterozygous knockout (FIR⁺/⁻) C57BL6 mice were generated. FIR complete knockout (FIR⁻/⁻) was embryonic lethal before E9.5; therefore, it is essential for embryogenesis. This strongly suggests that insufficiency of FIR is crucial for carcinogenesis. FIR⁺/⁻ mice exhibited prominent c-myc mRNA upregulation, particularly in the peripheral blood (PB), without any significant pathogenic phenotype. Furthermore, elevated FIRΔexon2/FIR mRNA expression was detected in human leukemia samples and cell lines. Because the single knockout of TP53 generates thymic lymphoma, FIR⁺/⁻TP53⁻/⁻ generated T-cell type acute lymphocytic/lymphoblastic leukemia (T-ALL) with increased organ or bone marrow invasion with poor prognosis. RNA-sequencing analysis of sorted thymic lymphoma cells revealed that the Notch signaling pathway was activated significantly in FIR⁺/⁻TP53⁻/⁻ compared with that in FIR⁺/⁺TP53⁻/⁻ mice. Notch1 mRNA expression in sorted thymic lymphoma cells was confirmed using qRT-PCR. In addition, flow cytometry revealed that c-myc mRNA was negatively correlated with FIR but positively correlated with Notch1 in sorted T-ALL/thymic lymphoma cells. Moreover, the knockdown of TP53 or c-myc using siRNA decreased Notch1 expression in cancer cells. In addition, an adenovirus vector encoding FIRΔexon2 cDNA increased bleomycin-induced DNA damage. Taken together, these data suggest that the altered expression of FIRΔexon2 increased Notch1 at least partially by activating c-Myc via a TP53-independent pathway. In conclusion, the alternative splicing of FIR, which generates FIRΔexon2, may contribute to both colorectal carcinogenesis and leukemogenesis.
Keywords: FBP interacting repressor (FIR); P53; T-ALL; haplo-insufficiency; leukemia; splicing variant.
Conflict of interest statement
We have no potential conflicts of interest to disclose.
Figures




References
-
- Duncan R, Bazar L, Michelotti G, Tomonaga T, Krutzsch H, Avigan M, Levens D. A sequence-specific, single-strand binding protein activates the far upstream element of c-myc and defines a new DNA-binding motif. Genes Dev. 1994;8:465–80. - PubMed
-
- Bazar L, Meighen D, Harris V, Duncan R, Levens D, Avigan M. Targeted melting and binding of a DNA regulatory element by a transactivator of c-myc. J Biol Chem. 1995;270:8241–8. - PubMed
-
- Avigan MI, Strober B, Levens D. A far upstream element stimulates c-myc expression in undifferentiated leukemia cells. J Biol Chem. 1990;265:18538–45. - PubMed
-
- Liu J, He L, Collins I, Ge H, Libutti D, Li J, Egly JM, Levens D. The FBP interacting repressor targets TFIIH to inhibit activated transcription. Mol Cell. 2000;5:331–41. - PubMed
-
- Liu J, Akoulitchev S, Weber A, Ge H, Chuikov S, Libutti D, Wang XW, Conaway JW, Harris CC, Conaway RC, Reinberg D, Levens D. Defective interplay of activators and repressors with TFIH in xeroderma pigmentosum. Cell. 2001;104:353–63. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

