Species typing in dermal leishmaniasis
- PMID: 25672782
- PMCID: PMC4402951
- DOI: 10.1128/CMR.00104-14
Species typing in dermal leishmaniasis
Abstract
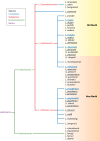
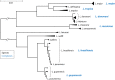
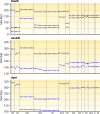
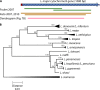
Leishmania is an infectious protozoan parasite related to African and American trypanosomes. All Leishmania species that are pathogenic to humans can cause dermal disease. When one is confronted with cutaneous leishmaniasis, identification of the causative species is relevant in both clinical and epidemiological studies, case management, and control. This review gives an overview of the currently existing and most used assays for species discrimination, with a critical appraisal of the limitations of each technique. The consensus taxonomy for the genus is outlined, including debatable species designations. Finally, a numerical literature analysis is presented that describes which methods are most used in various countries and regions in the world, and for which purposes.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures







References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

