La FAM fatale: USP9X in development and disease
- PMID: 25672900
- PMCID: PMC4427618
- DOI: 10.1007/s00018-015-1851-0
La FAM fatale: USP9X in development and disease
Abstract
Deubiquitylating enzymes (DUBs), act downstream of ubiquitylation. As such, these post-post-translational modifiers function as the final arbitrators of a protein substrate's ubiquitylation status, thus regulating its fate. In most instances, DUBs moderate the absolute level of a substrate, its locality or activity, rather than being an "all-or-none" phenomenon. Yet, disruption of this quantitative regulation can produce dramatic qualitative differences. The ubiquitin-specific protease 9X (USP9X/FAM) is a substrate-specific DUB, which displays an extraordinarily high level of sequence conservation from Drosophila to mammals. It is primarily the recent revelations of USP9X's pivotal role in human cancers, both as oncogene or tumour suppressor, in developmental disorders including intellectual disability, epilepsy, autism and developmental delay that has led to a subsequent re-examination of its molecular and cellular functions. Results from experimental animal models have implicated USP9X in neurodegeneration, including Parkinson's and Alzheimer's disease, as well as autoimmune diseases. In this review, we describe the current and accumulated knowledge on the molecular, cellular and developmental aspects of USP9X function within the context of the biological consequences during normal development and disease.
Figures
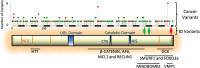
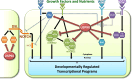
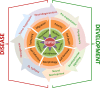
References
-
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. - PubMed
-
- Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. - PubMed
-
- Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. - PubMed
-
- Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004;1695:189–207. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

