Structure and receptor binding preferences of recombinant hemagglutinins from avian and human H6 and H10 influenza A virus subtypes
- PMID: 25673707
- PMCID: PMC4442393
- DOI: 10.1128/JVI.03456-14
Structure and receptor binding preferences of recombinant hemagglutinins from avian and human H6 and H10 influenza A virus subtypes
Abstract
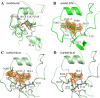
During 2013, three new avian influenza A virus subtypes, A(H7N9), A(H6N1), and A(H10N8), resulted in human infections. While the A(H7N9) virus resulted in a significant epidemic in China across 19 provinces and municipalities, both A(H6N1) and A(H10N8) viruses resulted in only a few human infections. This study focuses on the major surface glycoprotein hemagglutinins from both of these novel human viruses. The detailed structural and glycan microarray analyses presented here highlight the idea that both A(H6N1) and A(H10N8) virus hemagglutinins retain a strong avian receptor binding preference and thus currently pose a low risk for sustained human infections.
Importance: Human infections with zoonotic influenza virus subtypes continue to be a great public health concern. We report detailed structural analysis and glycan microarray data for recombinant hemagglutinins from A(H6N1) and A(H10N8) viruses, isolated from human infections in 2013, and compare them with hemagglutinins of avian origin. This is the first structural report of an H6 hemagglutinin, and our results should further the understanding of these viruses and provide useful information to aid in the continuous surveillance of these zoonotic influenza viruses.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures



Similar articles
-
A Portrait of the Sialyl Glycan Receptor Specificity of the H10 Influenza Virus Hemagglutinin-A Picture of an Avian Virus on the Verge of Becoming a Pandemic?Vaccines (Basel). 2017 Dec 13;5(4):51. doi: 10.3390/vaccines5040051. Vaccines (Basel). 2017. PMID: 29236069 Free PMC article. Review.
-
Unique Structural Features of Influenza Virus H15 Hemagglutinin.J Virol. 2017 May 26;91(12):e00046-17. doi: 10.1128/JVI.00046-17. Print 2017 Jun 15. J Virol. 2017. PMID: 28404848 Free PMC article.
-
A human-infecting H10N8 influenza virus retains a strong preference for avian-type receptors.Cell Host Microbe. 2015 Mar 11;17(3):377-384. doi: 10.1016/j.chom.2015.02.006. Cell Host Microbe. 2015. PMID: 25766296 Free PMC article.
-
Structural basis for preferential avian receptor binding by the human-infecting H10N8 avian influenza virus.Nat Commun. 2015 Jan 9;6:5600. doi: 10.1038/ncomms6600. Nat Commun. 2015. PMID: 25574798
-
Emergence in China of human disease due to avian influenza A(H10N8)--cause for concern?J Infect. 2014 Mar;68(3):205-15. doi: 10.1016/j.jinf.2013.12.014. Epub 2014 Jan 6. J Infect. 2014. PMID: 24406432 Review.
Cited by
-
A Portrait of the Sialyl Glycan Receptor Specificity of the H10 Influenza Virus Hemagglutinin-A Picture of an Avian Virus on the Verge of Becoming a Pandemic?Vaccines (Basel). 2017 Dec 13;5(4):51. doi: 10.3390/vaccines5040051. Vaccines (Basel). 2017. PMID: 29236069 Free PMC article. Review.
-
Bacterial glycosyltransferase-mediated cell-surface chemoenzymatic glycan modification.Nat Commun. 2019 Apr 17;10(1):1799. doi: 10.1038/s41467-019-09608-w. Nat Commun. 2019. PMID: 30996301 Free PMC article.
-
Structural insights into the design of novel anti-influenza therapies.Nat Struct Mol Biol. 2018 Feb;25(2):115-121. doi: 10.1038/s41594-018-0025-9. Epub 2018 Feb 2. Nat Struct Mol Biol. 2018. PMID: 29396418 Free PMC article. Review.
-
The Evolution, Spread and Global Threat of H6Nx Avian Influenza Viruses.Viruses. 2020 Jun 22;12(6):673. doi: 10.3390/v12060673. Viruses. 2020. PMID: 32580412 Free PMC article. Review.
-
Evaluation of the Biological Properties and Cross-Reactive Antibody Response to H10 Influenza Viruses in Ferrets.J Virol. 2017 Sep 12;91(19):e00895-17. doi: 10.1128/JVI.00895-17. Print 2017 Oct 1. J Virol. 2017. PMID: 28701401 Free PMC article.
References
-
- Fouchier RA, Munster V, Wallensten A, Bestebroer TM, Herfst S, Smith D, Rimmelzwaan GF, Olsen B, Osterhaus AD. 2005. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J Virol 79:2814–2822. doi:10.1128/JVI.79.5.2814-2822.2005. - DOI - PMC - PubMed
-
- Tong S, Li Y, Rivailler P, Conrardy C, Castillo DA, Chen LM, Recuenco S, Ellison JA, Davis CT, York IA, Turmelle AS, Moran D, Rogers S, Shi M, Tao Y, Weil MR, Tang K, Rowe LA, Sammons S, Xu X, Frace M, Lindblade KA, Cox NJ, Anderson LJ, Rupprecht CE, Donis RO. 2012. A distinct lineage of influenza A virus from bats. Proc Natl Acad Sci U S A 109:4269–4274. doi:10.1073/pnas.1116200109. - DOI - PMC - PubMed
-
- Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, Yang H, Chen X, Recuenco S, Gomez J, Chen LM, Johnson A, Tao Y, Dreyfus C, Yu W, McBride R, Carney PJ, Gilbert AT, Chang J, Guo Z, Davis CT, Paulson JC, Stevens J, Rupprecht CE, Holmes EC, Wilson IA, Donis RO. 2013. New World bats harbor diverse influenza A viruses. PLoS Pathog 9:e1003657. doi:10.1371/journal.ppat.1003657. - DOI - PMC - PubMed
-
- Thompson MG, Shay DK, Zhou H, Bridges CB, Cheng PY, Burns E, Bresee JS, Cox NJ. 2010. Estimates of deaths associated with seasonal influenza—United States, 1976-2007. MMWR Morb Mortal Wkly Rep 59:1057–1062. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

