Role of PGC-1α during acute exercise-induced autophagy and mitophagy in skeletal muscle
- PMID: 25673772
- PMCID: PMC4420796
- DOI: 10.1152/ajpcell.00380.2014
Role of PGC-1α during acute exercise-induced autophagy and mitophagy in skeletal muscle
Abstract
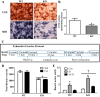
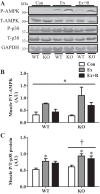
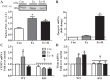
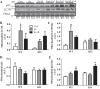
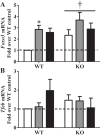
Regular exercise leads to systemic metabolic benefits, which require remodeling of energy resources in skeletal muscle. During acute exercise, the increase in energy demands initiate mitochondrial biogenesis, orchestrated by the transcriptional coactivator peroxisome proliferator-activated receptor-γ coactivator-1α (PGC-1α). Much less is known about the degradation of mitochondria following exercise, although new evidence implicates a cellular recycling mechanism, autophagy/mitophagy, in exercise-induced adaptations. How mitophagy is activated and what role PGC-1α plays in this process during exercise have yet to be evaluated. Thus we investigated autophagy/mitophagy in muscle immediately following an acute bout of exercise or 90 min following exercise in wild-type (WT) and PGC-1α knockout (KO) animals. Deletion of PGC-1α resulted in a 40% decrease in mitochondrial content, as well as a 25% decline in running performance, which was accompanied by severe acidosis in KO animals, indicating metabolic distress. Exercise induced significant increases in gene transcripts of various mitochondrial (e.g., cytochrome oxidase subunit IV and mitochondrial transcription factor A) and autophagy-related (e.g., p62 and light chain 3) genes in WT, but not KO, animals. Exercise also resulted in enhanced targeting of mitochondria for mitophagy, as well as increased autophagy and mitophagy flux, in WT animals. This effect was attenuated in the absence of PGC-1α. We also identified Niemann-Pick C1, a transmembrane protein involved in lysosomal lipid trafficking, as a target of PGC-1α that is induced with exercise. These results suggest that mitochondrial turnover is increased following exercise and that this effect is at least in part coordinated by PGC-1α.
Keywords: Niemann-Pick C1; biogenesis; endurance; mitochondria; physical activity.
Copyright © 2015 the American Physiological Society.
Figures



Similar articles
-
Exercise induces TFEB expression and activity in skeletal muscle in a PGC-1α-dependent manner.Am J Physiol Cell Physiol. 2018 Jan 1;314(1):C62-C72. doi: 10.1152/ajpcell.00162.2017. Epub 2017 Oct 18. Am J Physiol Cell Physiol. 2018. PMID: 29046293 Free PMC article.
-
PGC-1alpha plays a functional role in exercise-induced mitochondrial biogenesis and angiogenesis but not fiber-type transformation in mouse skeletal muscle.Am J Physiol Cell Physiol. 2010 Mar;298(3):C572-9. doi: 10.1152/ajpcell.00481.2009. Epub 2009 Dec 23. Am J Physiol Cell Physiol. 2010. PMID: 20032509 Free PMC article.
-
Autophagy activation, not peroxisome proliferator-activated receptor γ coactivator 1α, may mediate exercise-induced improvements in glucose handling during diet-induced obesity.Exp Physiol. 2017 Sep 1;102(9):1194-1207. doi: 10.1113/EP086406. Epub 2017 Jul 22. Exp Physiol. 2017. PMID: 28639297
-
PGC-1alpha-mediated adaptations in skeletal muscle.Pflugers Arch. 2010 Jun;460(1):153-62. doi: 10.1007/s00424-010-0834-0. Epub 2010 Apr 19. Pflugers Arch. 2010. PMID: 20401754 Review.
-
Deacetylation of PGC-1α by SIRT1: importance for skeletal muscle function and exercise-induced mitochondrial biogenesis.Appl Physiol Nutr Metab. 2011 Oct;36(5):589-97. doi: 10.1139/h11-070. Epub 2011 Sep 2. Appl Physiol Nutr Metab. 2011. PMID: 21888529 Review.
Cited by
-
Autophagy and ALS: mechanistic insights and therapeutic implications.Autophagy. 2022 Feb;18(2):254-282. doi: 10.1080/15548627.2021.1926656. Epub 2021 May 31. Autophagy. 2022. PMID: 34057020 Free PMC article. Review.
-
PGC-1β maintains mitochondrial metabolism and restrains inflammatory gene expression.Sci Rep. 2022 Sep 26;12(1):16028. doi: 10.1038/s41598-022-20215-6. Sci Rep. 2022. PMID: 36163487 Free PMC article.
-
Mouse skeletal muscle adaptations to different durations of treadmill exercise after the cessation of FOLFOX chemotherapy.Front Physiol. 2023 Nov 1;14:1283674. doi: 10.3389/fphys.2023.1283674. eCollection 2023. Front Physiol. 2023. PMID: 38028800 Free PMC article.
-
The mediating role of inflammaging between mitochondrial dysfunction and sarcopenia in aging: a review.Am J Clin Exp Immunol. 2023 Dec 15;12(6):109-126. eCollection 2023. Am J Clin Exp Immunol. 2023. PMID: 38187366 Free PMC article. Review.
-
Contractile activity attenuates autophagy suppression and reverses mitochondrial defects in skeletal muscle cells.Autophagy. 2018;14(11):1886-1897. doi: 10.1080/15548627.2018.1491488. Epub 2018 Aug 4. Autophagy. 2018. PMID: 30078345 Free PMC article.
References
-
- Adhihetty PJ, Uguccioni G, Leick L, Hidalgo J, Pilegaard H, Hood DA. The role of PGC-1α on mitochondrial function and apoptotic susceptibility in muscle. Am J Physiol Cell Physiol 297: C217–C225, 2009. - PubMed
-
- Baar K, Wende AR, Jones TE, Marison M, Nolte LA, Chen M, Kelly DP, Holloszy JO. Adaptations of skeletal muscle to exercise: rapid increase in the transcriptional coactivator PGC-1. FASEB J 16: 1879–1886, 2002. - PubMed
-
- Geisler S, Holmström KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol 12: 119–131, 2010. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

