ALTERED MERISTEM PROGRAM1 suppresses ectopic stem cell niche formation in the shoot apical meristem in a largely cytokinin-independent manner
- PMID: 25673776
- PMCID: PMC4378165
- DOI: 10.1104/pp.114.254623
ALTERED MERISTEM PROGRAM1 suppresses ectopic stem cell niche formation in the shoot apical meristem in a largely cytokinin-independent manner
Abstract
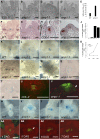
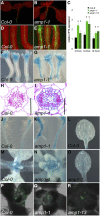
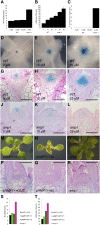
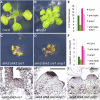
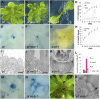
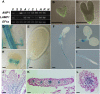
Plants are able to reiteratively form new organs in an environmentally adaptive manner during postembryonic development. Organ formation in plants is dependent on stem cell niches (SCNs), which are located in the so-called meristems. Meristems show a functional zonation along the apical-basal axis and the radial axis. Shoot apical meristems of higher plants are dome-like structures, which contain a central SCN that consists of an apical stem cell pool and an underlying organizing center. Organ primordia are formed in the circular peripheral zone (PZ) from stem cell descendants in which differentiation programs are activated. One mechanism to keep this radial symmetry integrated is that the existing SCN actively suppresses stem cell identity in the PZ. However, how this lateral inhibition system works at the molecular level is far from understood. Here, we show that a defect in the putative carboxypeptidase ALTERED MERISTEM PROGRAM1 (AMP1) causes the formation of extra SCNs in the presence of an intact primary shoot apical meristem, which at least partially contributes to the enhanced shoot meristem size and leaf initiation rate found in the mutant. This defect appears to be neither a specific consequence of the altered cytokinin levels in amp1 nor directly mediated by the WUSCHEL/CLAVATA feedback loop. De novo formation of supernumerary stem cell pools was further enhanced in plants mutated in both AMP1 and its paralog LIKE AMP1, indicating that they exhibit partially overlapping roles to suppress SCN respecification in the PZ.
© 2015 American Society of Plant Biologists. All Rights Reserved.
Figures


References
-
- Aichinger E, Kornet N, Friedrich T, Laux T (2012) Plant stem cell niches. Annu Rev Plant Biol 63: 615–636 - PubMed
-
- Barton MK. (2010) Twenty years on: the inner workings of the shoot apical meristem, a developmental dynamo. Dev Biol 341: 95–113 - PubMed
-
- Bennett T, Sieberer T, Willett B, Booker J, Luschnig C, Leyser O (2006) The Arabidopsis MAX pathway controls shoot branching by regulating auxin transport. Curr Biol 16: 553–563 - PubMed
-
- Bollman KM, Aukerman MJ, Park MY, Hunter C, Berardini TZ, Poethig RS (2003) HASTY, the Arabidopsis ortholog of exportin 5/MSN5, regulates phase change and morphogenesis. Development 130: 1493–1504 - PubMed
-
- Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R (2000) Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289: 617–619 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

