Non-coding RNA: what is functional and what is junk?
- PMID: 25674102
- PMCID: PMC4306305
- DOI: 10.3389/fgene.2015.00002
Non-coding RNA: what is functional and what is junk?
Abstract
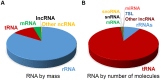
The genomes of large multicellular eukaryotes are mostly comprised of non-protein coding DNA. Although there has been much agreement that a small fraction of these genomes has important biological functions, there has been much debate as to whether the rest contributes to development and/or homeostasis. Much of the speculation has centered on the genomic regions that are transcribed into RNA at some low level. Unfortunately these RNAs have been arbitrarily assigned various names, such as "intergenic RNA," "long non-coding RNAs" etc., which have led to some confusion in the field. Many researchers believe that these transcripts represent a vast, unchartered world of functional non-coding RNAs (ncRNAs), simply because they exist. However, there are reasons to question this Panglossian view because it ignores our current understanding of how evolution shapes eukaryotic genomes and how the gene expression machinery works in eukaryotic cells. Although there are undoubtedly many more functional ncRNAs yet to be discovered and characterized, it is also likely that many of these transcripts are simply junk. Here, we discuss how to determine whether any given ncRNA has a function. Importantly, we advocate that in the absence of any such data, the appropriate null hypothesis is that the RNA in question is junk.
Keywords: Junk DNA; Junk RNA; evolution; genome biology; non-coding RNA.
Figures
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

