Novel inhibitors of cholesterol degradation in Mycobacterium tuberculosis reveal how the bacterium's metabolism is constrained by the intracellular environment
- PMID: 25675247
- PMCID: PMC4335503
- DOI: 10.1371/journal.ppat.1004679
Novel inhibitors of cholesterol degradation in Mycobacterium tuberculosis reveal how the bacterium's metabolism is constrained by the intracellular environment
Abstract
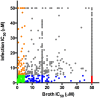
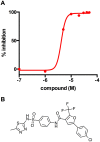
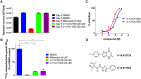
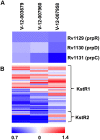
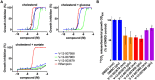
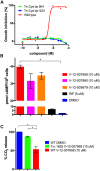
Mycobacterium tuberculosis (Mtb) relies on a specialized set of metabolic pathways to support growth in macrophages. By conducting an extensive, unbiased chemical screen to identify small molecules that inhibit Mtb metabolism within macrophages, we identified a significant number of novel compounds that limit Mtb growth in macrophages and in medium containing cholesterol as the principle carbon source. Based on this observation, we developed a chemical-rescue strategy to identify compounds that target metabolic enzymes involved in cholesterol metabolism. This approach identified two compounds that inhibit the HsaAB enzyme complex, which is required for complete degradation of the cholesterol A/B rings. The strategy also identified an inhibitor of PrpC, the 2-methylcitrate synthase, which is required for assimilation of cholesterol-derived propionyl-CoA into the TCA cycle. These chemical probes represent new classes of inhibitors with novel modes of action, and target metabolic pathways required to support growth of Mtb in its host cell. The screen also revealed a structurally-diverse set of compounds that target additional stage(s) of cholesterol utilization. Mutants resistant to this class of compounds are defective in the bacterial adenylate cyclase Rv1625/Cya. These data implicate cyclic-AMP (cAMP) in regulating cholesterol utilization in Mtb, and are consistent with published reports indicating that propionate metabolism is regulated by cAMP levels. Intriguingly, reversal of the cholesterol-dependent growth inhibition caused by this subset of compounds could be achieved by supplementing the media with acetate, but not with glucose, indicating that Mtb is subject to a unique form of metabolic constraint induced by the presence of cholesterol.
Conflict of interest statement
The authors CM EP DDD TW CPL are employees of Vertex Pharmaceuticals Incorporated. This does not alter our adherence to all PLOS policies on sharing data and materials.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

