Visualization of a radical B12 enzyme with its G-protein chaperone
- PMID: 25675500
- PMCID: PMC4345561
- DOI: 10.1073/pnas.1419582112
Visualization of a radical B12 enzyme with its G-protein chaperone
Abstract
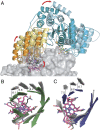
G-protein metallochaperones ensure fidelity during cofactor assembly for a variety of metalloproteins, including adenosylcobalamin (AdoCbl)-dependent methylmalonyl-CoA mutase and hydrogenase, and thus have both medical and biofuel development applications. Here, we present crystal structures of IcmF, a natural fusion protein of AdoCbl-dependent isobutyryl-CoA mutase and its corresponding G-protein chaperone, which reveal the molecular architecture of a G-protein metallochaperone in complex with its target protein. These structures show that conserved G-protein elements become ordered upon target protein association, creating the molecular pathways that both sense and report on the cofactor loading state. Structures determined of both apo- and holo-forms of IcmF depict both open and closed enzyme states, in which the cofactor-binding domain is alternatively positioned for cofactor loading and for catalysis. Notably, the G protein moves as a unit with the cofactor-binding domain, providing a visualization of how a chaperone assists in the sequestering of a precious cofactor inside an enzyme active site.
Keywords: G protein; crystallography; metallochaperone; metallocofactor delivery; vitamin B12.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
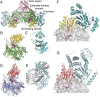
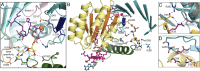

References
-
- Tottey S, Harvie DR, Robinson NJ. Understanding how cells allocate metals using metal sensors and metallochaperones. Acc Chem Res. 2005;38(10):775–783. - PubMed
-
- Finney LA, O’Halloran TV. Transition metal speciation in the cell: Insights from the chemistry of metal ion receptors. Science. 2003;300(5621):931–936. - PubMed
-
- Leipe DD, Wolf YI, Koonin EV, Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol. 2002;317(1):41–72. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

