Gene expression defines natural changes in mammalian lifespan
- PMID: 25677554
- PMCID: PMC4406664
- DOI: 10.1111/acel.12283
Gene expression defines natural changes in mammalian lifespan
Abstract
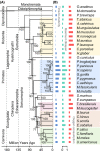
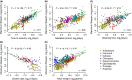
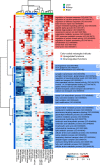
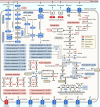
Mammals differ more than 100-fold in maximum lifespan, which can be altered in either direction during evolution, but the molecular basis for natural changes in longevity is not understood. Divergent evolution of mammals also led to extensive changes in gene expression within and between lineages. To understand the relationship between lifespan and variation in gene expression, we carried out RNA-seq-based gene expression analyses of liver, kidney, and brain of 33 diverse species of mammals. Our analysis uncovered parallel evolution of gene expression and lifespan, as well as the associated life-history traits, and identified the processes and pathways involved. These findings provide direct insights into how nature reversibly adjusts lifespan and other traits during adaptive radiation of lineages.
Keywords: aging; gene expression; life-history traits; lifespan; mammals.
© 2015 The Authors. Aging Cell published by the Anatomical Society and John Wiley & Sons Ltd.
Figures



References
-
- Beldade P, Brakefield PM, Long AD. Contribution of Distal-less to quantitative variation in butterfly eyespots. Nature. 2002;415:315–318. - PubMed
-
- Blomberg SP, Garland T, Jr, Ives AR. Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution. 2003;57:717–745. - PubMed
-
- Brawand D, Soumillon M, Necsulea A, Julien P, Csardi G, Harrigan P, Weier M, Liechti A, Aximu-Petri A, Kircher M, Albert FW, Zeller U, Khaitovich P, Grutzner F, Bergmann S, Nielsen R, Paabo S, Kaessmann H. The evolution of gene expression levels in mammalian organs. Nature. 2011;478:343–348. - PubMed
-
- Burbea Z, Nakhoul F, Rosenberg S, Zoabi R, Skorecki K, Hochberg I, Miller-Lotan R, Benchetrit S, Weissgarten J, Knecht A, Tovbin D, Levy NS, Levy AP. Role of haptoglobin phenotype in end-stage kidney disease. Nephron Exp. Nephrol. 2004;97:e71–e76. - PubMed
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

