Rare variants at 16p11.2 are associated with common variable immunodeficiency
- PMID: 25678086
- PMCID: PMC4461447
- DOI: 10.1016/j.jaci.2014.12.1939
Rare variants at 16p11.2 are associated with common variable immunodeficiency
Abstract
Background: Common variable immunodeficiency (CVID) is characterized clinically by inadequate quantity and quality of serum immunoglobulins with increased susceptibility to infections, resulting in significant morbidity and mortality. Only a few genes have been uncovered, and the genetic background of CVID remains elusive to date for the majority of patients.
Objective: We sought to seek novel associations of genes and genetic variants with CVID.
Methods: We performed association analyses in a discovery cohort of 164 patients with CVID and 19,542 healthy control subjects genotyped on the Immuno BeadChip from Illumina platform; replication of findings was examined in an independent cohort of 135 patients with CVID and 2,066 healthy control subjects, followed by meta-analysis.
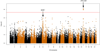
Results: We identified 11 single nucleotide polymorphisms (SNPs) at the 16p11.2 locus associated with CVID at a genome-wide significant level in the discovery cohort. The most significant SNP, rs929867 (P = 6.21 × 10(-9)), is in the gene fused-in-sarcoma (FUS), with 4 other SNPs mapping to integrin CD11b (ITGAM). Results were confirmed in our replication cohort. Conditional association analysis suggests a single association signal at the 16p11.2 locus. A strong trend of association was also seen for 38 SNPs (P < 5 × 10(-5)) in the MHC region, supporting that this is a genuine CVID locus. Interestingly, we found that 80% of patients with the rare ITGAM variants have reduced switched memory B-cell counts.
Conclusion: We report a novel association of CVID with rare variants at the FUS/ITGAM (CD11b) locus on 16p11.2. The association signal is enriched for promoter/enhancer markers in the ITGAM gene. ITGAM encodes the integrin CD11b, a part of complement receptor 3, a novel candidate gene implicated here for the first time in the pathogenesis of CVID.
Keywords: ITGAM; Immunodeficiency; genome-wide association study; immunogenetics; rare variants.
Copyright © 2015 American Academy of Allergy, Asthma & Immunology. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Figures

References
-
- Cunningham-Rundles C, Bodian C. Common variable immunodeficiency: clinical and immunological features of 248 patients. Clin Immunol. 1999;92:34–48. - PubMed
-
- Wehr C, Kivioja T, Schmitt C, Ferry B, Witte T, Eren E, et al. The EUROclass trial: defining subgroups in common variable immunodeficiency. Blood. 2008;111:77–85. - PubMed
-
- Salzer U, Chapel HM, Webster AD, Pan-Hammarstrom Q, Schmitt-Graeff A, Schlesier M, et al. Mutations in TNFRSF13B encoding TACI are associated with common variable immunodeficiency in humans. Nat Genet. 2005;37:820–828. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

