Draft genome sequence of Halomonas lutea strain YIM 91125(T) (DSM 23508(T)) isolated from the alkaline Lake Ebinur in Northwest China
- PMID: 25678942
- PMCID: PMC4315136
- DOI: 10.1186/1944-3277-10-1
Draft genome sequence of Halomonas lutea strain YIM 91125(T) (DSM 23508(T)) isolated from the alkaline Lake Ebinur in Northwest China
Abstract
Species of the genus Halomonas are halophilic and their flexible adaption to changes of salinity and temperature brings considerable potential biotechnology applications, such as degradation of organic pollutants and enzyme production. The type strain Halomonas lutea YIM 91125(T) was isolated from a hypersaline lake in China. The genome of strain YIM 91125(T) becomes the twelfth species sequenced in Halomonas, and the thirteenth species sequenced in Halomonadaceae. We described the features of H. lutea YIM 91125(T), together with the high quality draft genome sequence and annotation of its type strain. The 4,533,090 bp long genome of strain YIM 91125(T) with its 4,284 protein-coding and 84 RNA genes is a part of Genomic Encyclopedia of Type Strains, Phase I: the one thousand microbial genomes (KMG-I) project. From the viewpoint of comparative genomics, H. lutea has a larger genome size and more specific genes, which indicated acquisition of function bringing better adaption to its environment. DDH analysis demonstrated that H. lutea is a distinctive species, and halophilic features and nitrogen metabolism related genes were discovered in its genome.
Keywords: Aerobic; Chemoorganotrophic; Gram-negative; Halomonas lutea; Lake Ebinur; Moderately halophilic.
Figures
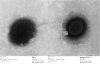


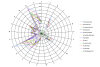
References
-
- Franzmann PD, Wehmeyer U, Stackebrandt E. Halomonadaceae fam. nov., a New Family of the Class Proteobacteria to Accommodate the Genera Halomonas and Deleya. Syst Appl Microbiol. 1988;11:16–19. doi: 10.1016/S0723-2020(88)80043-2. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

