Evolution of genome size and complexity in the rhabdoviridae
- PMID: 25679389
- PMCID: PMC4334499
- DOI: 10.1371/journal.ppat.1004664
Evolution of genome size and complexity in the rhabdoviridae
Abstract
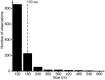
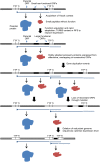
RNA viruses exhibit substantial structural, ecological and genomic diversity. However, genome size in RNA viruses is likely limited by a high mutation rate, resulting in the evolution of various mechanisms to increase complexity while minimising genome expansion. Here we conduct a large-scale analysis of the genome sequences of 99 animal rhabdoviruses, including 45 genomes which we determined de novo, to identify patterns of genome expansion and the evolution of genome complexity. All but seven of the rhabdoviruses clustered into 17 well-supported monophyletic groups, of which eight corresponded to established genera, seven were assigned as new genera, and two were taxonomically ambiguous. We show that the acquisition and loss of new genes appears to have been a central theme of rhabdovirus evolution, and has been associated with the appearance of alternative, overlapping and consecutive ORFs within the major structural protein genes, and the insertion and loss of additional ORFs in each gene junction in a clade-specific manner. Changes in the lengths of gene junctions accounted for as much as 48.5% of the variation in genome size from the smallest to the largest genome, and the frequency with which new ORFs were observed increased in the 3' to 5' direction along the genome. We also identify several new families of accessory genes encoded in these regions, and show that non-canonical expression strategies involving TURBS-like termination-reinitiation, ribosomal frame-shifts and leaky ribosomal scanning appear to be common. We conclude that rhabdoviruses have an unusual capacity for genomic plasticity that may be linked to their discontinuous transcription strategy from the negative-sense single-stranded RNA genome, and propose a model that accounts for the regular occurrence of genome expansion and contraction throughout the evolution of the Rhabdoviridae.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


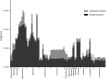


References
-
- Holmes EC (2009) The Evolution and Emergence of RNA Viruses. Oxford: Oxford University Press.
-
- King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors (2012) Virus Taxonomy. Classification and Nomenclature of Viruses Ninth report of the International Committee on Taxonomy of Viruses. London: Elsevier Academic Press.
-
- Steinhauer DA, Domingo E, Holland JJ (1992) Lack of evidence for proofreading mechanisms associated with an RNA virus polymerase. Gene 122: 281–288. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

