Integrative multi-omics module network inference with Lemon-Tree
- PMID: 25679508
- PMCID: PMC4332478
- DOI: 10.1371/journal.pcbi.1003983
Integrative multi-omics module network inference with Lemon-Tree
Abstract
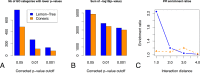
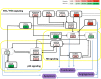
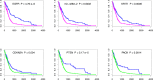
Module network inference is an established statistical method to reconstruct co-expression modules and their upstream regulatory programs from integrated multi-omics datasets measuring the activity levels of various cellular components across different individuals, experimental conditions or time points of a dynamic process. We have developed Lemon-Tree, an open-source, platform-independent, modular, extensible software package implementing state-of-the-art ensemble methods for module network inference. We benchmarked Lemon-Tree using large-scale tumor datasets and showed that Lemon-Tree algorithms compare favorably with state-of-the-art module network inference software. We also analyzed a large dataset of somatic copy-number alterations and gene expression levels measured in glioblastoma samples from The Cancer Genome Atlas and found that Lemon-Tree correctly identifies known glioblastoma oncogenes and tumor suppressors as master regulators in the inferred module network. Novel candidate driver genes predicted by Lemon-Tree were validated using tumor pathway and survival analyses. Lemon-Tree is available from http://lemon-tree.googlecode.com under the GNU General Public License version 2.0.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, et al. (2005) Integrative genomic analyses identify mitf as a lineage survival oncogene amplified in malignant melanoma. Nature 436: 117–122. - PubMed
-
- Segal E, Shapira M, Regev A, Pe’er D, Botstein D, et al. (2003) Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat Genet 34: 166–167. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

