Transoceanic spreading of pathogenic strains of Vibrio parahaemolyticus with distinctive genetic signatures in the recA gene
- PMID: 25679989
- PMCID: PMC4334540
- DOI: 10.1371/journal.pone.0117485
Transoceanic spreading of pathogenic strains of Vibrio parahaemolyticus with distinctive genetic signatures in the recA gene
Abstract
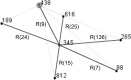
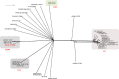
Vibrio parahaemolyticus is an important human pathogen whose transmission is associated with the consumption of contaminated seafood. Consistent multilocus sequence typing for V. parahaemolyticus has shown difficulties in the amplification of the recA gene by PCR associated with a lack of amplification or a larger PCR product than expected. In one strain (090-96, Peru, 1996), the produced PCR product was determined to be composed of two recA fragments derived from different Vibrio species. To better understand this phenomenon, we sequenced the whole genome of this strain. The hybrid recA gene was found to be the result of a fragmentation of the original lineage-specific recA gene resulting from a DNA insertion of approximately 30 kb in length. This insert had a G+C content of 38.8%, lower than that of the average G+C content of V. parahaemolyticus (45.2%), and contained 19 ORFs, including a complete recA gene. This new acquired recA gene deviated 24% in sequence from the original recA and was distantly related to recA genes from bacteria of the Vibrionaceae family. The reconstruction of the original recA gene (recA3) identified the precursor as belonging to ST189, a sequence type reported previously only in Asian countries. The identification of this singular genetic feature in strains from Asia reveals new evidence for genetic connectivity between V. parahaemolyticus populations at both sides of the Pacific Ocean that, in addition to the previously described pandemic clone, supports the existence of a recurrent transoceanic spreading of pathogenic V. parahaemolyticus with the corresponding potential risk of pandemic expansion.
Conflict of interest statement
Figures


References
-
- Turner JW, Paranjpye RN, Landis ED, Biryukov SV, Gonzalez-Escalona N, et al. (2013) Population structure of clinical and environmental Vibrio parahaemolyticus from the Pacific Northwest coast of the United States. PLoS One 8: e55726 PONE-D-12-32492 [pii]. 10.1371/journal.pone.0055726 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

