Secretome of Trichoderma interacting with maize roots: role in induced systemic resistance
- PMID: 25681119
- PMCID: PMC4390251
- DOI: 10.1074/mcp.M114.046607
Secretome of Trichoderma interacting with maize roots: role in induced systemic resistance
Abstract
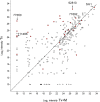
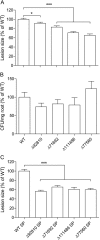
Trichoderma virens is a biocontrol agent used in agriculture to antagonize pathogens of crop plants. In addition to direct mycoparasitism of soil-borne fungal pathogens, T. virens interacts with roots. This interaction induces systemic resistance (ISR), which reduces disease in above-ground parts of the plant. In the molecular dialog between fungus and plant leading to ISR, proteins secreted by T. virens provide signals. Only a few such proteins have been characterized previously. To study the secretome, proteins were characterized from hydroponic culture systems with T. virens alone or with maize seedlings, and combined with a bioassay for ISR in maize leaves infected by the pathogen Cochliobolus heterostrophus. The secreted protein fraction from coculture of maize roots and T. virens (Tv+M) was found to have a higher ISR activity than from T. virens grown alone (Tv). A total of 280 fungal proteins were identified, 66 showing significant differences in abundance between the two conditions: 32 were higher in Tv+M and 34 were higher in Tv. Among the 34 found in higher abundance in Tv and negatively regulated by roots were 13 SSCPs (small, secreted, cysteine rich proteins), known to be important in the molecular dialog between plants and fungi. The role of four SSCPs in ISR was studied by gene knockout. All four knockout lines showed better ISR activity than WT without affecting colonization of maize roots. Furthermore, the secreted protein fraction from each of the mutant lines showed improved ISR activity compared with WT. These SSCPs, apparently, act as negative effectors reducing the defense levels in the plant and may be important for the fine tuning of ISR by Trichoderma. The down-regulation of SSCPs in interaction with plant roots implies a revision of the current model for the Trichoderma-plant symbiosis and its induction of resistance to pathogens.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




Similar articles
-
Host-specific transcriptomic pattern of Trichoderma virens during interaction with maize or tomato roots.BMC Genomics. 2015 Jan 22;16(1):8. doi: 10.1186/s12864-014-1208-3. BMC Genomics. 2015. PMID: 25608961 Free PMC article.
-
Sm2, a paralog of the Trichoderma cerato-platanin elicitor Sm1, is also highly important for plant protection conferred by the fungal-root interaction of Trichoderma with maize.BMC Microbiol. 2015 Jan 16;15(1):2. doi: 10.1186/s12866-014-0333-0. BMC Microbiol. 2015. PMID: 25591782 Free PMC article.
-
Plant-derived sucrose is a key element in the symbiotic association between Trichoderma virens and maize plants.Plant Physiol. 2009 Oct;151(2):792-808. doi: 10.1104/pp.109.141291. Epub 2009 Aug 12. Plant Physiol. 2009. PMID: 19675155 Free PMC article.
-
Trichoderma: The Current Status of Its Application in Agriculture for the Biocontrol of Fungal Phytopathogens and Stimulation of Plant Growth.Int J Mol Sci. 2022 Feb 19;23(4):2329. doi: 10.3390/ijms23042329. Int J Mol Sci. 2022. PMID: 35216444 Free PMC article. Review.
-
Endemic plants harbour specific Trichoderma communities with an exceptional potential for biocontrol of phytopathogens.J Biotechnol. 2016 Oct 10;235:162-70. doi: 10.1016/j.jbiotec.2016.03.049. Epub 2016 Mar 30. J Biotechnol. 2016. PMID: 27039271 Review.
Cited by
-
Trichoderma virens and Pseudomonas chlororaphis Differentially Regulate Maize Resistance to Anthracnose Leaf Blight and Insect Herbivores When Grown in Sterile versus Non-Sterile Soils.Plants (Basel). 2024 Apr 30;13(9):1240. doi: 10.3390/plants13091240. Plants (Basel). 2024. PMID: 38732455 Free PMC article.
-
Identification of miRNAs Involved in Maize-Induced Systemic Resistance Primed by Trichoderma harzianum T28 against Cochliobolus heterostrophus.J Fungi (Basel). 2023 Feb 20;9(2):278. doi: 10.3390/jof9020278. J Fungi (Basel). 2023. PMID: 36836392 Free PMC article.
-
The impact of microbes in the orchestration of plants' resistance to biotic stress: a disease management approach.Appl Microbiol Biotechnol. 2019 Jan;103(1):9-25. doi: 10.1007/s00253-018-9433-3. Epub 2018 Oct 12. Appl Microbiol Biotechnol. 2019. PMID: 30315353 Free PMC article. Review.
-
Early Transcriptome Response of Trichoderma virens to Colonization of Maize Roots.Front Fungal Biol. 2021 Aug 25;2:718557. doi: 10.3389/ffunb.2021.718557. eCollection 2021. Front Fungal Biol. 2021. PMID: 37744095 Free PMC article.
-
Target highlights in CASP14: Analysis of models by structure providers.Proteins. 2021 Dec;89(12):1647-1672. doi: 10.1002/prot.26247. Epub 2021 Oct 10. Proteins. 2021. PMID: 34561912 Free PMC article.
References
-
- Brotman Y., Kapuganti J. G., Viterbo A. (2010) Trichoderma. Curr. Biol. 20, R390–R391 - PubMed
-
- Lorito M., Woo S. L., Harman G. E., Monte E. (2010) Translational research on Trichoderma: from 'omics to the field. Annu. Rev. Phytopathol. 48, 395–417 - PubMed
-
- Harman G. E., Howell C. R., Viterbo A., Chet I., Lorito M. (2004) Trichoderma species-opportunistic, avirulent plant symbionts. Nat. Rev. Microbiol. 2, 43–56 - PubMed
-
- Benitez T., Rincón A. M., Limon M. C., Codón A. C. (2004) Biocontrol mechanisms of Trichoderma strains. Int. Microbiol. 7, 249–260 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

